| Revision as of 03:06, 22 February 2007 editExarion (talk | contribs)832 editsm Reverted 1 edit by 71.196.99.82 identified as vandalism to last revision by 128.187.0.164. TWINKLE← Previous edit | Revision as of 03:29, 22 February 2007 edit undo74.97.31.113 (talk)No edit summaryNext edit → | ||
| (One intermediate revision by the same user not shown) | |||
| Line 54: | Line 54: | ||
| Numerous polymerases can repicate DNA in eukaryotic cells. Currently, six families of polymerases (A, B, C, D, X, Y) have been discovered. At least four different types of DNA polymerases are involved in the replication of DNA in animal cells (POLA, POLG, POLD1 and POLE). POL1 functions by extending the primer in the 5' -> 3' direction and tightly associates with primase. However, it lacks the ability to proofread DNA. POLD1 has a proofreading ability and is able to replicate the entire length of a template only when associated with PCNA. POLE is able to replicate the entire length of a template in teh absence of PCNA and is able to proofread DNA while POLG replicates mitochondrial DNA via the D-Loop mechanism of DNA replicaton. All primers are removed by RNaseH1 and Flap Endonuclease I. The general mechanisms of DNA replication on the leading and lagging strand, however, are the same as to those found in prokaryotic cells. | Numerous polymerases can repicate DNA in eukaryotic cells. Currently, six families of polymerases (A, B, C, D, X, Y) have been discovered. At least four different types of DNA polymerases are involved in the replication of DNA in animal cells (POLA, POLG, POLD1 and POLE). POL1 functions by extending the primer in the 5' -> 3' direction and tightly associates with primase. However, it lacks the ability to proofread DNA. POLD1 has a proofreading ability and is able to replicate the entire length of a template only when associated with PCNA. POLE is able to replicate the entire length of a template in teh absence of PCNA and is able to proofread DNA while POLG replicates mitochondrial DNA via the D-Loop mechanism of DNA replicaton. All primers are removed by RNaseH1 and Flap Endonuclease I. The general mechanisms of DNA replication on the leading and lagging strand, however, are the same as to those found in prokaryotic cells. | ||
| ===Telomerase=== | |||
| A unique problem that occurs during the replication of linear chromosomes is chromosome shortening. Chromosome shortening occurs when the primer at the 5' end of the lagging strand is degraded. Because DNA polymerase cannot add new nucleotides to the 5' end of DNA (there is no place for a new primer), the ends would shorten after each round of replication. However, in most replicating cells a small amount of telomerase is present, and this enzyme extends the ends so that the chromosomes so that this problem does not occur. This extension occurs when the telomerase enzyme binds to a section of DNA on the 3' end and extends it using the normal replication machinery. This then allows for a primer to bind so that the complementary strand can be extended by normal lagging strand synthesis. Finally, telomeres must be capped by a protein to prevent chromosomal instability. | |||
| == References == | == References == | ||
Revision as of 03:29, 22 February 2007
| This article needs attention from an expert in Biology. Please add a reason or a talk parameter to this template to explain the issue with the article. WikiProject Biology may be able to help recruit an expert. |
| This article does not cite any sources. Please help improve this article by adding citations to reliable sources. Unsourced material may be challenged and removed. Find sources: "DNA replication" – news · newspapers · books · scholar · JSTOR (November 2006) (Learn how and when to remove this message) |

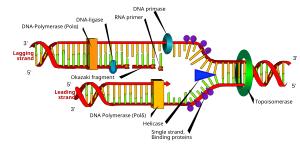
DNA replication or DNA synthesis is the process of copying a double-stranded DNA molecule. This process is paramount to all life as we know it and the general mechanisms of DNA replication is conserved among prokaryotic and eukaryotic organisms. DNA replication involves copying the genetic material and passing it on to daughter cells, therefore the process is important in continuation of life.
A DNA molecule is a long polymer built from nucelotides. Both strands in DNA run anti-parallel to eachother and are complementary to eachother. One of the DNA strands is built in the 5' → 3' direction, while the complementary strand is built in the 3' → 5' direction (5' and 3' each mark one end of a strand). The nucleotides which make up DNA pair with each other and form base pairs such as A::T and C:::G (two dots between A and T indicate that they are bound by two hydrogen bonds, and three dots between G and C indicate the presence of three hydrogen bonds). This means that the strand running in the 5'→ 3' direction will have base A that will pair with base T on the opposite strand running in 3'→ 5' direction.
Since DNA strands are antiparallel and complementary, each strand can serve as a template for the reproduction of the opposite strand. The template strand is preserved as a whole piece and the new strand is assembled from nucleotide triphosphates. This process is called semiconservative replication. Ideally, the two resulting strands are identical, although in reality there are always errors, though proofreading and error-checking mechanisms exist to ensure a very high level of fidelity.
In a cell, DNA replicaton is obligatory prior to cell division. Prokaryotes persistently replicate their DNA. In eukaryotes, timings are highly regulated and this occurs during the S phase of the cell cycle, preceding mitosis or meiosis I. The process may also be performed in vitro using reconstituted or completely artificial components, in a process known as PCR.
DNA may be synthesized artificially, but this process is not fundamentally a replicative process, and only produces a single strand of DNA.
Introduction
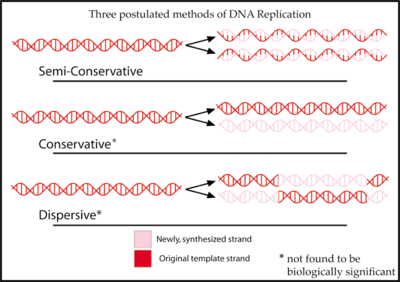
DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand. Since DNA strands are antiparallel and complementary, each strand can serve as a template for the reproduction of the opposite strand. The template strand is preserved as a whole piece and the new strand is assembled from nucleotide triphosphates. This process is called semiconservative replication. Ideally, the two resulting strands are identical, although in reality there are always errors, though proofreading and error-checking mechanisms exist to ensure a very high level of fidelity.
In a cell, this step is obligatory prior to cell division. Prokaryotes persistently replicate their DNA and creating a whole, new chromosome is a limiting step in cell division.The large size of eukaryotic chromosomes and the limits of nucleotide incorporation during DNA synthesis, make it necessary for multiple origins of replication to exist in order to complete replication in a reasonable period of time. However, it is clear that at a replication origin the strands of DNA must dissociate and unwind in order to allow access to DNA polymerase.
The process may also be performed in-vitro using reconstituted or completely artificial components, in a process known as PCR. DNA may be synthesized artificially, but this process is not fundamentally a replicative process, and only produces one strand of DNA.
Although the general mechanisms of DNA replication are conserved among all species, differences do exist between prokaryotic and eukaryotic DNA replication. The model organisms used to study DNA replicaton is E. coli (for prokaryotes) and yeast - specifically S. pombe and S. cerevisiae) for eukaryotes. The following explinations will take each case seperatly.
Prokaryotic DNA Replicaton
DNA replicaton in E. coli is bi-directional and originates at a single origin of replicaton (OriC). The initiation of replication is mediated by DnaA, a protein that binds to a region of the origin known as the DnaA box. In E. coli, there are 5 DnaA boxes, each of which contains a highly conserved 9bp consensous sequence 5' - TTATCCACA - 3'. Binding of DnaA to this region causes it to become negativly supercoiled. Following this, a region of OriC upstream of the DnaA boxes (known as DnaB boxes) become melted. There are three of these regions, and each are 13bp long, and AT rich (which obviously facilitates melting because less energy is required to break the two hydrogen bonds that form between A and T nucleotides). This region has the consensous sequence 5' - GATCTNTTNTTTT - 3. Melting of the DnaB boxes requires ATP (which is hydrolyzed by DnaA). Following melting, DnaA recuits a hexameric helicase (six DnaB proteins) to oppisate ends of the melted DNA. This is where the replication fork will form. Recruitment of helicase requires six DnaC proteins, each of which is attached to one subunit of helicase. Once this complex is formed, an additional five DnaA proteins bind to the original five DnaA proteins to form five DnaA dimers. DnaC is then released, and the prepriming complex is complete. In order for DNA replication to continue, SSB is needed to prevent the single strands of DNA from forming any secondary structures and to prevent them from reannealing, and DNA gyrase is needed to relieves the stress (by creating negative supercoils) created by the action of DnaB helicase. The unwinding of DNA by DnaB helicase allows for primase (DnaG) and RNA polymerase to prime each DNA template so that DNA synthesis can begin.
Once priming is complete, DNA polymerase III holoenzyme is loaded into the DNA and replication begins. The catalytic mechanism of DNA polymerase III involves the use of two metal ions in the active site, and a region in the active site that can discriminate between deoxynucleotides and ribonucleotides. The metal ions are general divalent cations that help the 3' OH initiate a nucleophilic attack onto the alpha phosphate of the deoxyribonucleotide and orient and stabalize the negatively charged triphosphate on the deoxyribonucleotide. Nucleophillic attack by the 3' OH on the alpha phosphate releases pyrophosphate, which is then subsequently hydrolyzed (by inorganic phosphatase) into two phophates. This hydrolysis drives DNA sysnthesis to completion.
Furthermore, DNA polymerase III must be able to distinguish between correctly paired bases and incorrectly paired bases. This is accomplished by distinguishing Watson-Crick base pairs through the use of an active site pocket that that is complementary in shape to the structure of correctly paired nucleotides. This pocket has a tyrosine residue that is able to form van der Waals interactions with the correctly paired nucleotide. In addition, dsDNA in the active site has a wider and shallower minor groove that permits the formation of hydrogen bonds with the third nitrogen of purine bases and the second oxygen of pyrimidine bases. Finally, the active site makes extensive hydrogen bonds with the DNA backbone. These interactions result in the DNA polymerase III closing around a correctly paired base. If a base is inserted and incorrectly paired, these interactions could not occur due to disruptions in hydrogen bonding and van der Waals interactions.
DNA is synthesized in one direction only, 5' -> 3'. However, one of the parent strands of DNA is 3' -> 5' and the other is 5' -> 3'. At the replication fork, one strand is synthesized continuously using the aformentioned mechanisms, while the other is synthesized in a series of short fragments known as Okazaki fragments. The 3' -> 5' strand is synthesized continuously, and is known as the leading strand. The RNA primers of Okazaki fragments are subsequently degraded by RNAseH and DNA Polymerase I, and the gap is filled with deoxyribonucleotides and sealed by the enzyme ligase.
Termination of DNA replication in E. coli is completed through the use of termination sequences and the Tus protein. These sequences allow the two replicaton forks to pass through in only one direction, but not the other. However, these sequences are not required for termination of replication.
Regulation of DNA replication is achieved through several mechanisms. Mechanisms involve the ratio of ATP to ADP, of DnaA to the number of DnaA boxes and the hemimethylation and sequestering of OriC. The ratio of ATP to ADP indicates that the cell has reached a specific size and is ready to divide. This "signal" occurs becuase in a rich medium, the cell will grow quickly and will have a lot of excess DNA. Futhermore, DnaA binds equally well to ATP or ADP, and only the DnaA-ATP complex is able to initate replication. Thus, in a fast growing cell, there will be more DnaA-ATP than DnaA-ADP. Because the levels of DnaA are strictly regulated, and 5 DnaA-DnaA dimers are needed to initiate replication, the ratio of DnaA to the number of DnaA boxes in the cell is important. After DNA replicaton is complete, this number is halved, thus DNA replication cannot occur until the levels of DnaA protein increases. Finally, DNA is sequestered to a membrane binding protein called SeqA. This protein binds to hemimethylated GATC DNA sequences. This four bp sequences occurs 11 times in OriC, and newly synthesized DNA only has its parent strand methylated. DAM methyltrasferase methylates the newly synthesized strand of DNA ONLY if it is not bound to SeqA. The importance of hemimethylation is twofold. Firstly, OriC becomes inaccessable to DnaA, and secondly, DnaA binds better to fully methylated DNA than hemimethylated DNA.
Eukaryotic DNA Replicaton
You must add a |reason= parameter to this Cleanup template – replace it with {{Cleanup|section|reason=<Fill reason here>}}, or remove the Cleanup template.
Although the mechanisms of DNA synthesis in Eukaryotes and prokaryotes is similar, DNA replication in eukaryotes is much more complicated. Though DNA synthesis in prokaryotes such as E. coli is regulated, DNA replication is initiated before the end of the cell cycle. Eukaryotic cells can only initiate DNA replication at a specific point in the cell cycle, the beginning of S phase.
DNA replication in eukaryotes occurs only in the S phase of the cell cycle. However, pre-initiation occurs in G1. Due to the sheer size of chromosomes in eukaryotes, eukaryotic chromosomes contain multiple origins of replication. Some origins are well characterized, such as the autonomously replicating sequences (ARS) of yeast while other eukaryotic origins, particularly those in metazoa, can be found in spans of thousands of basepairs. However, the assembly and initiation of replicaton is similar in both the protozoa and metazoa.
The first step in DNA replication is the formation of the pre-initiation replication complex (the pre-RC). The formation of this complex occurs in two stages. The first stage requires that there is no CDK activity. This can only occur in early G1. The formation of the pre-RC is known as licensing, but a licensed pre-RC cannot initaite replicaton. Initiation of replication can only occur during the S-phase. Thus, the seperation of licensing and activation ensures that the origin can only fire once per cell cycle.
DNA replication in eukaryotes is not very well characterized. However, researchers believe that it begins with the binding of the origin recognition complex (ORC) to the origin. This complex is a hexamer of related proteins and remains bound to the origin, even after DNA replication occurs. Furthermore, ORC is the functional analogue of DnaA. Following the binding of ORC to the origin, Cdc6/Cdc18 and Cdt1 coordinate the loading of the MCM (minichromosome maitenance functions) complex to the origin by first binding to ORC and then binding to the MCM complex. The MCM complex is thought to be the major DNA helicase in eukaryotic organisms, and is a hexamer (mcm2-7). Once binding of MCM occurs, a fully licensed pre-RC exists.
Activation of the complex occurs in S-phase and requires Cdk2-Cyclin E and Ddk. The activation process begins with the addition of Mcm10 to the pre-RC, which displaces Cdt1. Following this, Ddk phosphorylates Mcm3-7, which activates the helicase. It is believed that ORC and Cdc6/18 are phosphorylated by Cdk2-Cyclin E. Ddk and the the Cdk complex then recruits another protein called Cdc45, which then recruits all of the DNA replicaton proteins to the replicaton fork. At this stage the origin fires and DNA synthesis begins.
Activation of a new round of replicaton is prevented through the actions of the cyclin dependant kinases and a protein known as geminin. Geminin binds to Cdt1 and sequesters it. It is a periodic protein that first appears in S-phase and is degraded in late M-phase, possibly through the action of the anaphase promoting complex (APC). In addition, phosphorylation of Cdc6/18 prevent it from binding to the ORC (thus inhibiting loading of the MCM complex) while the phosphorylation of ORC remains unclear. Cells in the G0 stage of the cell cycle are prevented from initiating a round of replication because the Mcm proteins are not expressed.
Numerous polymerases can repicate DNA in eukaryotic cells. Currently, six families of polymerases (A, B, C, D, X, Y) have been discovered. At least four different types of DNA polymerases are involved in the replication of DNA in animal cells (POLA, POLG, POLD1 and POLE). POL1 functions by extending the primer in the 5' -> 3' direction and tightly associates with primase. However, it lacks the ability to proofread DNA. POLD1 has a proofreading ability and is able to replicate the entire length of a template only when associated with PCNA. POLE is able to replicate the entire length of a template in teh absence of PCNA and is able to proofread DNA while POLG replicates mitochondrial DNA via the D-Loop mechanism of DNA replicaton. All primers are removed by RNaseH1 and Flap Endonuclease I. The general mechanisms of DNA replication on the leading and lagging strand, however, are the same as to those found in prokaryotic cells.
Telomerase
A unique problem that occurs during the replication of linear chromosomes is chromosome shortening. Chromosome shortening occurs when the primer at the 5' end of the lagging strand is degraded. Because DNA polymerase cannot add new nucleotides to the 5' end of DNA (there is no place for a new primer), the ends would shorten after each round of replication. However, in most replicating cells a small amount of telomerase is present, and this enzyme extends the ends so that the chromosomes so that this problem does not occur. This extension occurs when the telomerase enzyme binds to a section of DNA on the 3' end and extends it using the normal replication machinery. This then allows for a primer to bind so that the complementary strand can be extended by normal lagging strand synthesis. Finally, telomeres must be capped by a protein to prevent chromosomal instability.
References
- Voet and Voet. Biochemistry, Third Edition (2004). ISBN 0-471-19350-X. Wiley International Edition.
- Watson, Baker, Bell, Gann, Levine, Losick. Molecular Biology of the Gene, Fifth Edition (2003). ISBN 0-8053-4635-X. Pearson/Benjamin Cummings Publishing.
External links
- DNA Workshop
- WEHI-TV - DNA Replication video- Detailed DNA replication animation from different angles with description below.
- Breakfast of Champions Does Replication- creative primer on the process from the Science Creative Quarterly
- Template:McGrawHillAnimation
- Basic Polymerase Chain Reaction Protocol
- Animated Biology
- DNA makes DNA (Flash Animation)
- DNA replication info pageby George Kakaris, Biologist MSc in Applied Genetics and Biotechnology
| DNA replication (comparing prokaryotic to eukaryotic) | |||||||
|---|---|---|---|---|---|---|---|
| Initiation |
| ||||||
| Replication |
| ||||||
| Termination | |||||||