| Revision as of 21:31, 20 January 2013 editSom999 (talk | contribs)Extended confirmed users1,215 edits Reverted 12 edits by Cavann (talk): Referenced content removed without citing any reason. (TW)← Previous edit | Revision as of 21:34, 20 January 2013 edit undoCavann (talk | contribs)Extended confirmed users2,026 edits Undid revision 534059033 by Som999 (talk) All changes are explained, see edit summaries. --1,581k edit is fixing previously cited sources to ref name formatNext edit → | ||
| Line 5: | Line 5: | ||
| ==Central Asian and Uralic connection== | ==Central Asian and Uralic connection== | ||
| ] | ] | ||
| The question to what extent a gene flow from ] to ] has contributed to the current gene pool of the Turkish people, and what the role is in this of the 11th century invasion by ], has been the subject of several studies. A factor that makes it difficult to give reliable estimates, is the problem of distinguishing between the effects of different migratory episodes. |
The question to what extent a gene flow from ] to ] has contributed to the current gene pool of the Turkish people, and what the role is in this of the 11th century invasion by ], has been the subject of several studies. A factor that makes it difficult to give reliable estimates, is the problem of distinguishing between the effects of different migratory episodes. Several studies have concluded that the historical and indigenous Anatolian groups are the primary source of the present-day Turkish population.<ref name=antigens57/><ref name=Yardumian_et_al/><ref name="Hodoglugil_et_al2"/><ref name=eurostudy/><ref name=humangenetics112/><ref name=stanford/><ref name=euroasia/> Thus, although the Turks carried out an invasion with ] significance, including the introduction of the ] and ], the genetic significance from ] might have been slight.<ref name=eurostudy/><ref name=med_pops></ref> | ||
| Some of the ] originated from Central Asia and therefore are possibly related with ].<ref name=Keyser-Tracqui_et_al>Christine Keyser-Tracqui, Eric Crubézy, Bertrand Ludes, Nuclear and Mitochondrial DNA Analysis of a 2,000-Year-Old Necropolis in the Egyin Gol Valley of Mongolia, The American Journal of Human Genetics, Volume 73, Issue 2, August 2003, Pages 247-260, ISSN 0002-9297, 10.1086/377005</ref> A majority (89%) of the Xiongnu sequences can be classified as belonging to an ] ] and nearly 11% belong to ] ].<ref name=Keyser-Tracqui_et_al/> This finding indicates that the contacts between European and Asian populations were anterior to the Xiongnu culture,<ref name=Keyser-Tracqui_et_al/> and it confirms results reported for two samples from an early 3rd century B.C. ]-] population.<ref name=Clisson2002>{{cite journal | |||
| | author = Clisson, I.; Keyser, C.; Francfort, H. P.; Crubezy, E.; Samashev, Z.; Ludes, B. | | author = Clisson, I.; Keyser, C.; Francfort, H. P.; Crubezy, E.; Samashev, Z.; Ludes, B. | ||
| | year = 2002 | | year = 2002 | ||
| Line 17: | Line 19: | ||
| }}</ref> | }}</ref> | ||
| ⚫ | According to another archeological and genetic study in 2010, the paternal Y-chromosome R1a, which is considered as an Indo-European marker, was found in three skeletons in 2000-year-old elite ] cemetery in Northeast Asia, which supports Kurgan expansion hypothesis for the Indo-European expansion from the Volga steppe region.<ref>Kim et al. A western Eurasian male is found in 2000-year-old elite Xiongnu cemetery in Northeast Mongolia, Am J Phys Anthropol. 2010 Jul;142(3):429-40, quoted pg.2 "The Kurgan expansion hypothesis explains the IndoEuropean expansion from the Volga steppe region (Gimbutas, 1973; Mallory, 1989).The paternal Y-chromosome single nucleotide polymorphisms (Y-SNP) R1a1 is considered as an Indo-European marker, supporting Kurgan expansion hypothesis (Zerjal et al., 1999; Kharkov et al., 2004; Haak et al., 2008). Recent finding of R1a1 in the Krasnoyarsk area east of Siberia marks the eastward expansion of the early Indo-Europeans (Keyser-Tracqui et al., 2009). R1a1 was not found in Scytho-Siberian skeletons from the Seby¨stei site of Altai Republic or in Xiongnu skeletons from Egyin Gol of Mongolia (KeyserTracqui et al., 2009)." quoted p.10: ", paternal, maternal, and biparental genetic analyses were done on three Xiongnu tombs of Northeast Mongolia 2,000 years ago. We showed for the first time that an Indo-European with paternal R1a1 and maternal U2e1 was present in the Xiongnu Empire of ancient Mongolia"</ref> As the R1a was found in Xiongnu people<ref>Kim et al. A western Eurasian male is found in 2000-year-old elite Xiongnu cemetery in Northeast Mongolia, Am J Phys Anthropol. 2010 Jul;142(3):429-40</ref> and the present-day people of Central Asia<ref>Xue et al. Male Demography in East Asia: A North–South Contrast in Human Population Expansion Times, Genetics. 2006April; 172(4): 2431–2439. doi: 10.1534/genetics.105.054270</ref> Analysis of skeletal remains from sites attributed to the Xiongnu provides an identification of dolichocephalic Mongoloid, ethnically distinct from neighboring populations in present-day Mongolia.<ref>Fu ren da xue (Beijing, China), S.V.D. Research Institute, Society of the Divine Word - 2003 </ref> | ||
| Some of these ancient specimens may be of a possible Turkic origin .<ref name=touchette/><ref name=Richards2000>{{cite journal | |||
| | author = Martin Richards, Vincent Macaulay, Eileen Hickey, Emilce Vega, Bryan Sykes, Valentina Guida, Chiara Rengo, Daniele Sellitto, Fulvio Cruciani, Toomas Kivisild, Richard Villems, Mark Thomas, Serge Rychkov, Oksana Rychkov, Yuri Rychkov, Mukaddes Gölge, Dimitar Dimitrov, Emmeline Hill11, Dan Bradley, Valentino Romano, Francesco Calì, Giuseppe Vona, Andrew Demaine, Surinder Papiha, Costas Triantaphyllidis, Gheorghe Stefanescu, Jiři Hatina, Michele Belledi, Anna Di Rienzo, Andrea Novelletto, Ariella Oppenheim, Søren Nørby, Nadia Al-Zaheri, Silvana Santachiara-Benerecetti, Rosaria Scozzari, Antonio Torroni, and Hans-Jürgen Bandelt | |||
| | title = Tracing European founder lineages in the Near Eastern mtDNA pool | |||
| | journal = American Journal of Human Genetics | |||
| | year = 2000 | |||
| | month = November | |||
| | volume = 67 | |||
| | issue = 5 | |||
| | pages = 1251–1276 | |||
| | url = http://www.stats.gla.ac.uk/~vincent/founder2000/ | |||
| | doi = 10.1016/S0002-9297(07)62954-1 | |||
| | pmid = 11032788 | |||
| | pmc = 1288566 | |||
| ⚫ | |||
| According to a different genetic research on 75 individuals from various parts of ], Mergen et al revealed that genetic structure of the mtDNAs in the ] population bears similarities to Turkic Central Asian populations. The neighbour-joining tree built from segment I sequences for Turkish and the other populations (French, Bulgarian, British, Finland, Greek, German, Kazakhs, Uighurs and Kirghiz) indicated two poles. Turkic Central Asian populations, Turkish population and British population formed one pole, and European populations formed the other, which revealed Turkish population bears more similarities to ] population and ].<ref>Hatice Mergen et al. Mitochondrial DNA sequence variation in the Anatolian Peninsula (Turkey), Journal of Genetics, Vol. 83, No.1, April 2004, article p.46 and fig.4 or http://www.ncbi.nlm.nih.gov/pubmed/15240908</ref> | According to a different genetic research on 75 individuals from various parts of ], Mergen et al revealed that genetic structure of the mtDNAs in the ] population bears similarities to Turkic Central Asian populations. The neighbour-joining tree built from segment I sequences for Turkish and the other populations (French, Bulgarian, British, Finland, Greek, German, Kazakhs, Uighurs and Kirghiz) indicated two poles. Turkic Central Asian populations, Turkish population and British population formed one pole, and European populations formed the other, which revealed Turkish population bears more similarities to ] population and ].<ref>Hatice Mergen et al. Mitochondrial DNA sequence variation in the Anatolian Peninsula (Turkey), Journal of Genetics, Vol. 83, No.1, April 2004, article p.46 and fig.4 or http://www.ncbi.nlm.nih.gov/pubmed/15240908</ref> | ||
| Nevertheless, today's Turkish people are more closely related with the Balkan populations than to the ] populations,<ref name=METU></ref><ref></ref> and a study looking into allele frequencies suggested that there was a lack of genetic relationship between the Mongols and the Turks, despite the historical relationship of their languages (The Turks and Germans were equally distant to all three Mongolian populations).<ref></ref> In addition, another study looking into HLA genes allele distributions indicated that Anatolians did not significantly differ from other Mediterranean populations.<ref name=med_pops></ref> Multiple studies suggested an ] ] dominance-driven ] model to explain the adoption of ] by ] indigenous inhabitants.<ref name=Yardumian_et_al/><ref name=euroasia/> | |||
| ==Haplogroup distributions in Turkish people== | ==Haplogroup distributions in Turkish people== | ||
Revision as of 21:34, 20 January 2013
| This article or section possibly contains synthesis of material that does not verifiably mention or relate to the main topic. Relevant discussion may be found on the talk page. (May 2010) (Learn how and when to remove this message) |
In population genetics the question has been debated whether the modern Turkish population is significantly related to other Turkic peoples, or whether they are rather derived from indigenous populations of Anatolia which were culturally assimilated during the Middle Ages. The contribution of the Central Asian genetics to the modern Turkish people has been debated and become the subject of several studies. As a result, several studies have concluded that the historical (pre-Islamic) and indigenous Anatolian groups are the primary source of the present-day Turkish population, in addition to in addition to neighboring peoples, such as Balkan peoples, and central Asian Turkic people.
Central Asian and Uralic connection

The question to what extent a gene flow from Central Asia to Anatolia has contributed to the current gene pool of the Turkish people, and what the role is in this of the 11th century invasion by Oghuz Turks, has been the subject of several studies. A factor that makes it difficult to give reliable estimates, is the problem of distinguishing between the effects of different migratory episodes. Several studies have concluded that the historical and indigenous Anatolian groups are the primary source of the present-day Turkish population. Thus, although the Turks carried out an invasion with cultural significance, including the introduction of the Turkish language and Islam, the genetic significance from Central Asia might have been slight.
Some of the Turkic peoples originated from Central Asia and therefore are possibly related with Xiongnu. A majority (89%) of the Xiongnu sequences can be classified as belonging to an Asian haplogroups and nearly 11% belong to European haplogroups. This finding indicates that the contacts between European and Asian populations were anterior to the Xiongnu culture, and it confirms results reported for two samples from an early 3rd century B.C. Scytho-Siberian population.
According to another archeological and genetic study in 2010, the paternal Y-chromosome R1a, which is considered as an Indo-European marker, was found in three skeletons in 2000-year-old elite Xiongnu cemetery in Northeast Asia, which supports Kurgan expansion hypothesis for the Indo-European expansion from the Volga steppe region. As the R1a was found in Xiongnu people and the present-day people of Central Asia Analysis of skeletal remains from sites attributed to the Xiongnu provides an identification of dolichocephalic Mongoloid, ethnically distinct from neighboring populations in present-day Mongolia.
According to a different genetic research on 75 individuals from various parts of Turkey, Mergen et al revealed that genetic structure of the mtDNAs in the Turkish population bears similarities to Turkic Central Asian populations. The neighbour-joining tree built from segment I sequences for Turkish and the other populations (French, Bulgarian, British, Finland, Greek, German, Kazakhs, Uighurs and Kirghiz) indicated two poles. Turkic Central Asian populations, Turkish population and British population formed one pole, and European populations formed the other, which revealed Turkish population bears more similarities to Turkic Central Asian population and British people.
Nevertheless, today's Turkish people are more closely related with the Balkan populations than to the Central Asian populations, and a study looking into allele frequencies suggested that there was a lack of genetic relationship between the Mongols and the Turks, despite the historical relationship of their languages (The Turks and Germans were equally distant to all three Mongolian populations). In addition, another study looking into HLA genes allele distributions indicated that Anatolians did not significantly differ from other Mediterranean populations. Multiple studies suggested an elite cultural dominance-driven linguistic replacement model to explain the adoption of Turkish language by Anatolian indigenous inhabitants.
Haplogroup distributions in Turkish people
According to Cinnioglu et al., (2004) there are many Y-DNA haplogroups present in Turkey. The majority haplogroups are primarily shared with Middle Eastern, Caucasian, and European populations such as haplogroups E3b, G, J, I, R1a, R1b, K and T which form 78.5% from the Turkish Gene pool (without R1b, K, and which notably occur elsewhere, it is 59.3%) and contrast with a smaller share of haplogroups related to Central Asia (N and Q)- 5.7% (but it rises to 36% if K, R1a, R1b and L- which infrequently occur in Central Asia, but are notable in many other Western Turkic groups), India H, R2 - 1.5% and Africa A, E3*, E3a - 1%. Some of the percentages identified were:
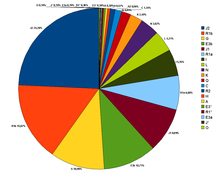
- J2=24% - J2 (M172) Typical of populations of Caucasus, the Near East, Southeast Europe, Southwest Asia with a moderate distribution through much of Central Asia, South Asia, and North Africa.
- R1b=14.7% -Typical of Western Europeans, Eurasian People, and typical of Uyghurs in the Central Asia
- G=10.9% - Typical of people from the Caucasus and to a lesser extent the Middle East.
- E1b1b1=10.7% - Typical of people from the Mediterranean
- J1=9% - Typical amongst people from the Arabian Peninsula and Dagestan (ranging from 3% from Turks around Konya to 12% in Kurds).
- R1a=6.9% - Typical of Central Asian, Caucasus, Altaic people, Eastern Europeans and Indo-Aryan people.
- I=5.3% - Typical of Central Europeans, Western Caucasian and Balkan populations.
- K=4.5% - Typical of Asian populations and Caucasian populations.
- L=4.2% - Typical of Indian Subcontinent and Khorasan populations.
- N=3.8% - Typical of Uralic, Siberian and Altaic populations.
- T=2.5% - Typical of Mediterranean, Middle Eastern, Northeast African and South Asian populations
- Q=1.9% - Typical of Northern Altaic populations.
Further research on Turkish Y-DNA groups
A study from Turkey by Gokcumen (2008) took into account oral histories and historical records. They went to four settlements in Central Anatolia and did not do a random selection from a group of university students like many other studies. Accordingly here are the results:
1) At an Afshar village whose oral stories tell they come from Central Asia they found that 57% come from haplogroup L, 13% from haplogroup Q, 3% from haplogroup N thus indicating that the L haplogroups in Turkey are of Central Asian heritage rather than Indian, although these Central Asians would have gotten the L markers from the Indians from the beginning. These Asian groups add up to 73% in this village. Furthermore 10% of these Afshars were E3a and E3b. Only 13% were J2a, the most common haplogroup in Turkey.
2) An older Turkish village center that did not receive much migration was about 25% N and 25% J2a with 3% G and close to 30% of some sort of R1 but mostly R1b.
Other Studies

In 2001, Benedetto et al revealed that Central Asian genetic contribution to the current Anatolian mtDNA gene pool was estimated as roughly 30%, by comparing the populations of Mediterranean Europe, and Turkic-speaking people of Central Asia. In 2003, Cinnioğlu et al. made a research of Y-DNA including the samples from eight regions of Turkey, without classifying the ethnicity of the people, which indicated that high resolution SNP analysis totally provides evidence of a detectable weak signal (<9%) of gene flow from Central Asia. In 2006, Berkman concluded that the true Central Asian contribution to Anatolia for both males and females were assumed to be 22%, with respect to the Balkans. In 2011 Aram Yardumian and Theodore G. Schurr published their study "Who Are the Anatolian Turks? A Reappraisal of the Anthropological Genetic Evidence." They revealed the impossibility of long-term, and continuing genetic contacts between Anatolia and Siberia, and confirmed the presence of significant mitochondrial DNA and Y-chromosome divergence between this regions, with minimal admixture. The research confirms also the lack of mass migration and suggested that it was irregular punctuated migration events that engendered large-scale shifts in language and culture among Anatolia's diverse autochthonous inhabitants. According to a 2012 study on ethnic Turks of Turkey, Hodoğlugil revealed that there is a significant overlap between Turks and Middle Easterners and a relationship with Europeans and South and Central Asians when Kyrgyz samples are genotyped and analysed. It displays a genetic ancestry for the Turks of 45% Middle Eastern, 40% European and 15% Central Asian. However, the Turkish genetic structure is unique, and there is an admixture of Turkish people reflecting the population migration patterns.
Furthermore, and maybe more so, in a study called 'The Genetics of Modern Assyrians and their Relationship to Other People of the Middle East' Dr. Joel J. Elias, a Professor at University of California School of Medicine at San Francisco conducted a genetic test about Turkish people. The aim was to discover the origins of the peoples of the Middle East presently, and over time. He found that "In spite of the complex history of the Middle East and the great number of internal group migrations revealed by history, as well as the mosaic of cultures and languages, the region is relatively homogeneous" in relation to the people that speak different languages throughout the region including Iranian and Kurdish (Indo-European), Turkish (Turkic) and Arabic, Assyrian and Aramaic (Semitic). A group of Armenian scientists conducted a study about the origins of the Turkish people in relation to Armenians. Savak Avagian; director of Armenia's bone marrow bank found that “Turks and Armenians were the two societies throughout the world that were genetically close to each other. Kurds are also in same genetic pool”.
See also
- Demographics of Turkey
- History of the Turkish people
- Archaeogenetics of the Near East
- Genetic history of Europe
References and notes
- ^ (2001) HLA alleles and haplotypes in the Turkish population: relatedness to Kurds, Armenians and other Mediterraneans Tissue Antigens 57 (4), 308–317
- ^ Yardumian, A., & Schurr, T. G. (2011). Who Are the Anatolian Turks?. Anthropology & Archeology Of Eurasia, 50(1), 6-42. doi:10.2753/AAE1061-1959500101
- ^ Hodoğlugil, U., & Mahley, R. W. (2012). Turkish Population Structure and Genetic Ancestry Reveal Relatedness among Eurasian Populations. Annals Of Human Genetics, 76(2), 128-141. doi:10.1111/j.1469-1809.2011.00701.x
- ^ Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language.
- ^ Testing hypotheses of language replacement in the neighbouring Caucasus: evidence from the Y-chromosome. Human genetics. 2003, vol. 112, no3, pp. 255–261. ISSN 0340-6717
- ^ Excavating Y-chromosome haplotype strata in Anatolia. Department of Genetics, Stanford University School of Medicine, 300 Pasteur Drive, Stanford, CA 94305-5120, USA.
- ^ The Eurasian Heartland: A continental perspective on Y-chromosome diversity.Proceedings of the National Academy of Sciences of the United States of America, Vol. 98, No. 18 (Aug. 28, 2001), pp. 10244–10249.
- Alu insertion polymorphisms in the Balkans and the origins of the Aromuns. Annals of Human Genetics.Volume 68 Issue 2 Page 120-127, March 2004.
- Mergen et al. Mitochondrial DNA sequence variation in the Anatolian Peninsula (Turkey), Journal of Genetics, Vol. 83, No.1, April 2004, P.46, Figure 4. http://www.ias.ac.in/jgenet/Vol83No1/39.pdf & http://www.ncbi.nlm.nih.gov/pubmed/15240908
- ^ Tissue Antigens Volume 60 Issue 2 Page 111-121, August(2002) Population genetic relationships between Mediterranean populations determined by HLA allele distribution and a historic perspective. Tissue Antigens 60 (2), 111–121
- ^ Christine Keyser-Tracqui, Eric Crubézy, Bertrand Ludes, Nuclear and Mitochondrial DNA Analysis of a 2,000-Year-Old Necropolis in the Egyin Gol Valley of Mongolia, The American Journal of Human Genetics, Volume 73, Issue 2, August 2003, Pages 247-260, ISSN 0002-9297, 10.1086/377005
- Clisson, I.; Keyser, C.; Francfort, H. P.; Crubezy, E.; Samashev, Z.; Ludes, B. (2002). "Genetic analysis of human remains from a double inhumation in a frozen kurgan in Kazakhstan". International Journal of Legal Medicine. 116 (5): 304–308. doi:10.1007/s00414-002-0295-x. PMID 12376844.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Kim et al. A western Eurasian male is found in 2000-year-old elite Xiongnu cemetery in Northeast Mongolia, Am J Phys Anthropol. 2010 Jul;142(3):429-40, quoted pg.2 "The Kurgan expansion hypothesis explains the IndoEuropean expansion from the Volga steppe region (Gimbutas, 1973; Mallory, 1989).The paternal Y-chromosome single nucleotide polymorphisms (Y-SNP) R1a1 is considered as an Indo-European marker, supporting Kurgan expansion hypothesis (Zerjal et al., 1999; Kharkov et al., 2004; Haak et al., 2008). Recent finding of R1a1 in the Krasnoyarsk area east of Siberia marks the eastward expansion of the early Indo-Europeans (Keyser-Tracqui et al., 2009). R1a1 was not found in Scytho-Siberian skeletons from the Seby¨stei site of Altai Republic or in Xiongnu skeletons from Egyin Gol of Mongolia (KeyserTracqui et al., 2009)." quoted p.10: ", paternal, maternal, and biparental genetic analyses were done on three Xiongnu tombs of Northeast Mongolia 2,000 years ago. We showed for the first time that an Indo-European with paternal R1a1 and maternal U2e1 was present in the Xiongnu Empire of ancient Mongolia"
- Kim et al. A western Eurasian male is found in 2000-year-old elite Xiongnu cemetery in Northeast Mongolia, Am J Phys Anthropol. 2010 Jul;142(3):429-40
- Xue et al. Male Demography in East Asia: A North–South Contrast in Human Population Expansion Times, Genetics. 2006April; 172(4): 2431–2439. doi: 10.1534/genetics.105.054270
- Fu ren da xue (Beijing, China), S.V.D. Research Institute, Society of the Divine Word - 2003
- Hatice Mergen et al. Mitochondrial DNA sequence variation in the Anatolian Peninsula (Turkey), Journal of Genetics, Vol. 83, No.1, April 2004, article p.46 and fig.4 Online Read or http://www.ncbi.nlm.nih.gov/pubmed/15240908
- Alu insertion polymorphisms and an assessment of the genetic contribution of Central Asia to Anatolia with respect to the Balkans. Department of Biological Sciences, Middle East Technical University, 06531 Ankara, Turkey. American Journal of Physical Anthropoly 2008 May;136(1):11-8.
- Alu insertion polymorphisms in the Balkans and the origins of the Aromuns. Annals of Human Genetics.Volume 68 Issue 2 Page 120-127, March 2004.
- Tissue Antigens. Volume 61 Issue 4 Page 292–299, April 2003. Genetic affinities among Mongol ethnic groups and their relationship to Turks
- ^ Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet (2004) 114 : 127–148, Springer-Verlag 2003
- Klyosov A.A. "The principal mystery in the relationship of Indo-European and Türkic linguistic families, and an attempt to solve it with the help of DNA genealogy: reflections of a non-linguist"//Proceedings of Russian Academy of DNA Genealogy (ISSN 1942-7484), Vol. 3, No 1, pp. 3 - 58
- Y Haplogroups of the World Online Edition
- Gokcumen O. et al (2008), Ethnohistorical and genetic survey of four Central Anatolian settlements, a dissertation/thesis
- Gokcumen O. Ethnohistorical and genetic survey of four central Anatolian settlements, dissertation, Univ. of Pennsylvania, 2008.
- Varzari et al. (2007) Population history of the Dniester-Carpathians: evidence from Alu markers. J Hum Genet. 2007;52(4):308-16.
- Online Reference Di Benedetto G, Ergüven A, Stenico M, Castrì L, Bertorelle G, Togan I, Barbujani G., DNA diversity and population admixture in Anatolia, Am J Phys Anthropol. 115(2):144-56, 2001. quoted: "The Turkic language was introduced in Anatolia at the start of this millennium, by nomadic Turkmen groups from Central Asia. Whether that cultural transition also had significant population-genetics consequences is not fully understood. Three nuclear microsatellite loci, the hypervariable region I of the mitochondrial genome, six microsatellite loci of the Y chromosome, and one Alu insertion (YAP) were amplified and typed in 118 individuals from four populations of Anatolia. For each locus, the number of chromosomes considered varied between 51-200. Genetic variation was large within samples, and much less so between them. The contribution of Central Asian genes to the current Anatolian gene pool was quantified using three different methods, considering for comparison populations of Mediterranean Europe, and Turkic-speaking populations of Central Asia. The most reliable estimates suggest roughly 30% Central Asian admixture for both mitochondrial and Y-chromosome loci. That (admittedly approximate) figure is compatible both with a substantial immigration accompanying the arrival of the Turkmen armies (which is not historically documented), and with continuous gene flow from Asia into Anatolia, at a rate of 1% for 40 generations. Because a military invasion is expected to more deeply affect the male gene pool, similar estimates of admixture for female- and male-transmitted traits are easier to reconcile with continuous migratory contacts between Anatolia and its Asian neighbors, perhaps facilitated by the disappearance of a linguistic barrier between them." [http://www.ncbi.nlm.nih.gov/pubmed/11385601 Online Reference
- "Excavating Y-chromosome haplotype strata in Anatolia." Human Genetics 114:2 (January 2004): pages 127-148. First published electronically on October 29, 2003. 523
- Ceren Berkman, Comparative Analyses for the Central Asian Contribution to Anatolian Gene Pool with Reference to Balkans, p.98, METU, Sep. 2006 quoted "Lower male than female contribution from Central Asia to Anatolia was obtained. The situation was explained by invoking the idea of homogenization between the males of the Balkans and Anatolia. Since females could not migrate alone, the true Central Asian contribution for both males and females were assumed to be 22%."
- Yardumian A, Schurr TG. 2011. Who are the Anatolian Turks? A reappraisal of the anthropological genetic evidence. Archeol Anthropol Eurasia 50(1): 6-43 (Summer).
- Uğur Hodoğlugil and Robert W. Mahley - Turkish Population Structure and Genetic Ancestry Reveal Relatedness among Eurasian Populations, Annals of Human Genetics, March 2012, Volume 76, Issue 2, pg. 128-141. Abstract "Turkey has experienced major population movements. Population structure and genetic relatedness of samples from three regions of Turkey, using over 500,000 SNP genotypes, were compared together with Human Genome Diversity Panel (HGDP) data. To obtain a more representative sampling from Central Asia, Kyrgyz samples (Bishkek, Kyrgyzstan) were genotyped and analysed. Principal component (PC) analysis reveals a significant overlap between Turks and Middle Easterners and a relationship with Europeans and South and Central Asians; however, the Turkish genetic structure is unique. FRAPPE, STRUCTURE, and phylogenetic analyses support the PC analysis depending upon the number of parental ancestry components chosen. For example, supervised STRUCTURE (K=3) illustrates a genetic ancestry for the Turks of 45% Middle Eastern (95% CI, 42-49), 40% European (95% CI, 36-44) and 15% Central Asian (95% CI, 13-16), whereas at K=4 the genetic ancestry of the Turks was 38% European (95% CI, 35-42), 35% Middle Eastern (95% CI, 33-38), 18% South Asian (95% CI, 16-19) and 9% Central Asian (95% CI, 7-11). PC analysis and FRAPPE/STRUCTURE results from three regions in Turkey (Aydin, Istanbul and Kayseri) were superimposed, without clear subpopulation structure, suggesting sample homogeneity. Thus, this study demonstrates admixture of Turkish people reflecting the population migration patterns."
- The Genetics of Modern Assyrians and their Relationship to Other People of the Middle East>
- ref name= Turks, Armenians share similar genes, say scientists (Hürriyet Daily News) >
http://en.wikipedia.org/Haplogroup_J2_%28Y-DNA%29
Categories: