This is an old revision of this page, as edited by SvartMan (talk | contribs) at 00:57, 10 October 2024 (→Uses: Removed section lacking in citations). The present address (URL) is a permanent link to this revision, which may differ significantly from the current revision.
Revision as of 00:57, 10 October 2024 by SvartMan (talk | contribs) (→Uses: Removed section lacking in citations)(diff) ← Previous revision | Latest revision (diff) | Newer revision → (diff)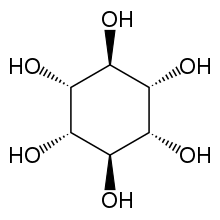
| |
| Names | |
|---|---|
| IUPAC name muco-Inositol | |
| Systematic IUPAC name (1R,2r,3S,4R,5r,6S)-Cyclohexane-1,2,3,4,5,6-hexol | |
| Identifiers | |
| CAS Number | |
| 3D model (JSmol) | |
| ChEBI | |
| ChemSpider | |
| ECHA InfoCard | 100.006.983 |
| IUPHAR/BPS | |
| UNII | |
InChI
| |
SMILES
| |
| Properties | |
| Chemical formula | C6H12O6 |
| Molar mass | 180.156 g·mol |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C , 100 kPa).
| |
Muco-inositol is one of nine stereo-isomers of inositol.
Nomenclature
Nomenclature is extremely important as it relates to muco-inositol. The utilization of this material in the neural system of the biological entity is totally dependent on the precise stereo-chemistry of this stereo-isomer. Unfortunately, the nomenclature has gone through a series of significant changes during the last thirty years. Only the literature subsequent to 1988 can be depended upon in this regard.
Muco-inositol (CAS 488-55-1) is a particular isomer of (and frequently confused with) the generic cyclohexane 1,2,3,4,5,6 hexol (CAS 87-89-8). This confusion should be avoided. The correct "chair" representation of muco-inositol is shown here. The numbering reflects the recommended 1988 numbering based on the fact that the isomer is typically phosphorylated at the hydroxyl group associated with the #1 carbon when used as the hydrated sodium receptor.
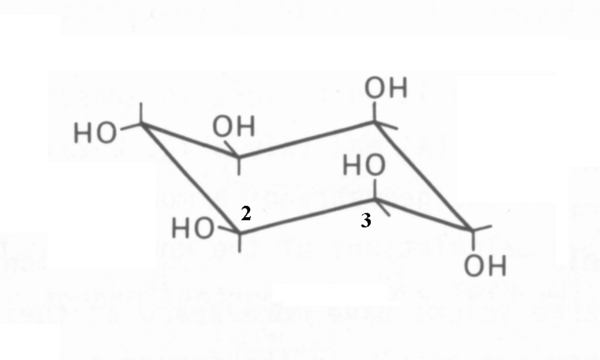
It is quite difficult to represent the critical stereo-graphic features of muco-inositol without employing the three-dimensional representation provided by the Jmol 3D images in the Chembox at upper right. The reason is that the distances between pairs of specific oxygen atoms are critically important to its operation as the active portion of the sodium channel sensory receptor. The values calculated using the Jmol script on this page will be used in this article in place of the preferred but unavailable measured values of these distances. There are many inaccurate Jmol representations of muco-inositol present on the internet. Please use caution and verify the accuracy of any other Jmol script used.
Detailed nomenclature
Note, the O3 and O4 atoms are both associated with axial hydroxyl groups pointed in opposite direction and separated by the single carbon-carbon bond of C3 and C4. The angles between the carbon-hydroxyl group bonds and the carbon-carbon bond are nominally 109.5 degrees.
The parameter of primary interest in gustation is the distance between the oxygen atoms of the pair O3 and O4 and the pair O4 and O5 in 3D space. This distance is defined as the d-value of the sensory receptor. This value cannot be obtained from the various Zig-Zag and Haworth representations commonly used in chemistry. Currently, the mean of the d-value of muco-inositol is between 3.3 Angstrom (0.33 nm) and 3.66 Angstrom (0.366 nm). The latter value is computed using the 3D structures found in the Jmol and Protein Data Bank libraries of Biochemistry. In both cases, another digit of precision is needed in the calculations to resolve this spread. Such precision is difficult to obtain in bond length measurements. The d-value of the O3 and O4 pair and the O4 and O5 pair are equal.
The d-value between the two oxygen atoms of a pair of hydroxyl groups is slightly different than the distance described in most of the work of Shallenberger and colleagues for the saccharides during the 1970s. They initially described the slightly longer distance between the hydrogen of one group and the oxygen of the other. They were the first to describe the dual hydrogen (or London) bonds between the two oxygens of a pair and a similar pair of oxygen atoms of a stimulant as the potential selection mechanism (first of two steps in the transduction process) in sweet or G-path gustatory sensing. The more detailed definition and comparison is shown in the figure.
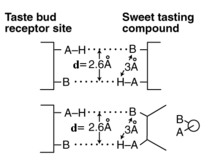
The d-value and ligand changes when other paths are selected. A more complete discussion of this mechanism is available.
Nomenclature of sodium ion in solution
To address the role of PtdIns in the first step of the two-step Na-path sensory transduction process, the conformation of the sodium ion in solution must be appreciated. It cannot exist as a free ion in solution. Upon solvation, the total molecule is ionized and the sodium-ion is immediately hydrated, involving coordination chemistry, to form Na(H2O)n where n varies but is most commonly six.
See also
- D-chiro-Inositol
- L-chiro-Inositol
- allo-Inositol
- cis-Inositol
- epi-Inositol
- Generic form of the phosphorylated inositol
- neo-Inositol
- scyllo-Inositol
References
- International Union of Pure and Applied Chemistry (2014). Nomenclature of Organic Chemistry: IUPAC Recommendations and Preferred Names 2013. The Royal Society of Chemistry. p. 1415. doi:10.1039/9781849733069. ISBN 978-0-85404-182-4.
- NC-IUB (Moss, G. ed.) (1988) Numbering of atoms in myo-inositol http://www.chem.qmul.ac.uk/iupac/cyclitol/myo.html
- Eggers, S. Acree, T. & Shallenberger, R. (2000) Sweetness chemoreception theory and sweetness transduction Food Chem vol 68(1), pp 45-49
- Shallenberger, R. & Acree, T. (1967) Molecular theory of sweet taste Nature vol 216, pp 480-482
- Fulton, J. (2012) http://neuronresearch.net/taste/pdf/Why%20only%20four%20gustaphores.pdf