This is the current revision of this page, as edited by DinosaursLoveExistence (talk | contribs) at 18:05, 29 August 2024 (→References: Category). The present address (URL) is a permanent link to this version.
Revision as of 18:05, 29 August 2024 by DinosaursLoveExistence (talk | contribs) (→References: Category)(diff) ← Previous revision | Latest revision (diff) | Newer revision → (diff)
| |
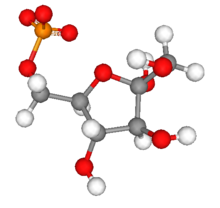
| |
| Names | |
|---|---|
| IUPAC name 6-O-Phosphono-α-D-fructofuranose | |
| Other names
β-D-fructose 6-phosphate, fructose 6-phosphate | |
| Identifiers | |
| CAS Number | |
| 3D model (JSmol) | |
| Abbreviations | F6P |
| ChEBI | |
| ChemSpider | |
| ECHA InfoCard | 100.010.360 |
| KEGG | |
| PubChem CID | |
| UNII | |
| CompTox Dashboard (EPA) | |
InChI
| |
SMILES
| |
| Properties | |
| Chemical formula | C6H13O9P |
| Molar mass | 260.14 g/mol |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C , 100 kPa).
| |
Fructose 6-phosphate (sometimes called the Neuberg ester) is a derivative of fructose, which has been phosphorylated at the 6-hydroxy group. It is one of several possible fructosephosphates. The β-D-form of this compound is very common in cells. The great majority of glucose is converted to fructose 6-phosphate upon entering a cell. Fructose is predominantly converted to fructose 1-phosphate by fructokinase following cellular import.
History
The name Neuberg ester comes from the German biochemist Carl Neuberg. In 1918, he found that the compound (later identified as fructose 6-phosphate) was produced by mild acid hydrolysis of fructose 2,6-bisphosphate.
In glycolysis
Fructose 6-phosphate lies within the glycolysis metabolic pathway and is produced by isomerisation of glucose 6-phosphate. It is in turn further phosphorylated to fructose-1,6-bisphosphate.
| α-D-glucose 6-phosphate | Phosphoglucose isomerase | α-D-fructose 6-phosphate | Phosphofructokinase-1 | α-D-fructose 1,6-bisphosphate | ||

|

|

| ||||
| ATP | ADP | |||||

|
| |||||
| Pi | H2O | |||||
| Phosphoglucose isomerase | Fructose bisphosphatase | |||||
Compound C00668 at KEGG Pathway Database. Enzyme 5.3.1.9 at KEGG Pathway Database. Compound C05345 at KEGG Pathway Database. Enzyme 2.7.1.11 at KEGG Pathway Database. Enzyme 3.1.3.11 at KEGG Pathway Database. Reaction at KEGG Pathway Database. Compound C05378 at KEGG Pathway Database.
Click on genes, proteins and metabolites below to link to respective articles.
[[File:

- The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
See also
| Glycolysis metabolic pathway | |
|---|---|
 ATP
ADP
ATP
ADP

 ATP
ADP
ATP
ADP


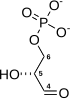
+ +
2 × Glyceraldehyde 3-phosphate 2 ×
Glyceraldehyde-3-phosphate  ADP
ATP
ADP
ATP
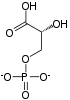
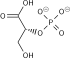
Phosphopyruvate  ADP
ATP
ADP
ATP
2 × Pyruvate 2 × |
References
- Berg, Jeremy M.; Tymoczko, Stryer (2002). Biochemistry (5th ed.). New York: W.H. Freeman and Company. ISBN 0-7167-3051-0.
- Nelson, D. L.; Cox, M. M. "Lehninger, Principles of Biochemistry" 3rd Ed. Worth Publishing: New York, 2000. ISBN 1-57259-153-6.
- Fruton, Joseph S. Proteins, Enzymes, Genes: The Interplay of Chemistry and Biology. Yale University Press: New Haven, 1999. p 292