This is an old revision of this page, as edited by Kjkolb (talk | contribs) at 05:59, 6 March 2014 ("amino acid residue" redirects here, wikified article (links, grammar) - not sure about choice to make additional sub-units, welcome changes by others). The present address (URL) is a permanent link to this revision, which may differ significantly from the current revision.
Revision as of 05:59, 6 March 2014 by Kjkolb (talk | contribs) ("amino acid residue" redirects here, wikified article (links, grammar) - not sure about choice to make additional sub-units, welcome changes by others)(diff) ← Previous revision | Latest revision (diff) | Newer revision → (diff)| This article needs additional citations for verification. Please help improve this article by adding citations to reliable sources. Unsourced material may be challenged and removed. Find sources: "Protein structure" – news · newspapers · books · scholar · JSTOR (March 2009) (Learn how and when to remove this message) |

Protein structure is the biomolecular structure of a protein molecule. Proteins are polymers – specifically polypeptides -sequences formed from various L-α-amino acids. Each unit of a protein is called an amino acid residue because it is the residue of every amino acid that forms the protein by losing a water molecule. By convention, a chain under 40 residues is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations, driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, and dual polarisation interferometry to determine the structure of proteins.
Protein structures range in size from tens to several thousand residues. By physical size, proteins are classified as nanoparticles, between 1–100 nm. Very large aggregates can be formed from protein subunits. For example, many thousand actin molecules assemble into a microfilament.
A protein may undergo reversible structural changes in performing its biological function. The alternative structures of the same protein are referred to as different conformations, and transitions between them are called conformational changes.
Levels of protein structure
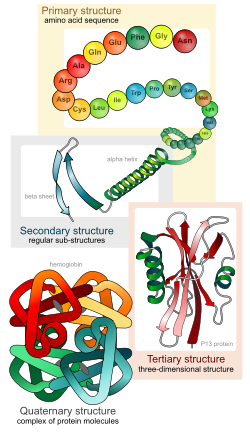
There are four distinct levels of protein structure.
Amino acid residues
Main article: Amino acid Main article: Proteinogenic amino acidEach α-amino acid consists of a backbone part that is present in all the amino acid types, and a side chain that is unique to each type of residue. An exception from this rule is proline. Because the carbon atom is bound to four different groups it is chiral, however only one of the isomers occur in biological proteins. Glycine however, is not chiral since its side chain is a hydrogen atom. A simple mnemonic for correct L-form is "CORN": when the Cα atom is viewed with the H in front, the residues read "CO-R-N" in a clockwise direction.
Primary structure
Main article: Protein primary structureThe primary structure of a protein refers to the linear sequence of amino acids in the polypeptide chain. The primary structure is held together by covalent bonds such as peptide bonds, which are made during the process of protein biosynthesis or translation. The two ends of the polypeptide chain are referred to as the carboxyl terminus (C-terminus) and the amino terminus (N-terminus) based on the nature of the free group on each extremity. Counting of residues always starts at the N-terminal end (NH2-group), which is the end where the amino group is not involved in a peptide bond. The primary structure of a protein is determined by the gene corresponding to the protein. A specific sequence of nucleotides in DNA is transcribed into mRNA, which is read by the ribosome in a process called translation. The sequence of amino acids was discovered by Frederick Sanger. The sequence of a protein is unique to that protein, and defines the structure and function of the protein. The sequence of a protein can be determined by methods such as Edman degradation or tandem mass spectrometry. Often, however, it is read directly from the sequence of the gene using the genetic code. We know that there are over 10,000 proteins in the human body which are composed of different arrangements of 20 types of amino acid residues. It is strictly recommended to use the word "amino acid residues" when discussing proteins because when a peptide bond is formed, a water molecule is lost, so proteins are made up of amino acid residues. Post-translational modification such as disulfide bond formation, phosphorylations and glycosylations are usually also considered a part of the primary structure, and cannot be read from the gene. For example, insulin is composed of 51 amino acids in 2 chains. One chain has 31 amino acids, and the other has 20 amino acids.
Secondary structure
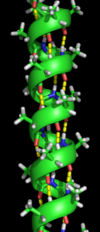
Secondary structure refers to highly regular local sub-structures. Two main types of secondary structure, the alpha helix and the beta strand or beta sheets, were suggested in 1951 by Linus Pauling and coworkers. These secondary structures are defined by patterns of hydrogen bonds between the main-chain peptide groups. They have a regular geometry, being constrained to specific values of the dihedral angles ψ and φ on the Ramachandran plot. Both the alpha helix and the beta sheet represent a way of saturating all the hydrogen bond donors and acceptors in the peptide backbone. Some parts of the protein are ordered but do not form any regular structures. They should not be confused with random coil, an unfolded polypeptide chain lacking any fixed three-dimensional structure. Several sequential secondary structures may form a "supersecondary unit".
Tertiary structure
Main article: Protein tertiary structureTertiary structure refers to the three-dimensional structure of a single, double, or triple bonded protein molecule. The alpha-helixes and beta pleated-sheets are folded into a compact globular structure. The folding is driven by the non-specific hydrophobic interactions, the burial of hydrophobic residues from water, but the structure is stable only when the parts of a protein domain are locked into place by specific tertiary interactions, such as salt bridges, hydrogen bonds, and the tight packing of side chains and disulfide bonds. The disulfide bonds are extremely rare in cytosolic proteins, since the cytosol (intracellular fluid) is generally a reducing environment.
Quaternary structure
Main article: Protein quaternary structureQuaternary structure is the three-dimensional structure of a multi-subunit protein and how the subunits fit together. In this context, the quaternary structure is stabilized by the same non-covalent interactions and disulfide bonds as the tertiary structure. Complexes of two or more polypeptides (i.e. multiple subunits) are called multimers. Specifically it would be called a dimer if it contains two subunits, a trimer if it contains three subunits, a tetramer if it contains four subunits, and a pentamer if it contains five subunits. The subunits are frequently related to one another by symmetry operations, such as a 2-fold axis in a dimer. Multimers made up of identical subunits are referred to with a prefix of "homo-" (e.g. a homotetramer) and those made up of different subunits are referred to with a prefix of "hetero-", for example, a heterotetramer, such as the two alpha and two beta chains of hemoglobin.
Domains, motifs, and folds in protein structure

Proteins are frequently described as consisting of several structural units. These units include domains, motifs, and folds. Despite the fact that there are about 100,000 different proteins expressed in eukaryotic systems, there are many fewer different domains, structural motifs and folds.
Structural domain
A structural domain is an element of the protein's overall structure that is self-stabilizing and often folds independently of the rest of the protein chain. Many domains are not unique to the protein products of one gene or one gene family but instead appear in a variety of proteins. Domains often are named and singled out because they figure prominently in the biological function of the protein they belong to; for example, the "calcium-binding domain of calmodulin". Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera proteins.
Structural and sequence motif
The structural and sequence motifs refer to short segments of protein three-dimensional structure or amino acid sequence that were found in a large number of different proteins.
Supersecondary structure
The supersecondary structure refers to a specific combination of secondary structure elements, such as beta-alpha-beta units or a helix-turn-helix motif. Some of them may be also referred to as structural motifs.
Protein fold
A protein fold refers to the general protein architecture, like a helix bundle, beta-barrel, Rossman fold or different "folds" provided in the Structural Classification of Proteins database.
Protein folding
Main article: Protein foldingAn unfolded polypeptide folds into its characteristic three-dimensional structure from a random coil.
Protein structure determination
Around 90% of the protein structures available in the Protein Data Bank have been determined by X-ray crystallography. This method allows one to measure the three-dimensional (3-D) density distribution of electrons in the protein, in the crystallized state, and thereby infer the 3-D coordinates of all the atoms to be determined to a certain resolution. Roughly 9% of the known protein structures have been obtained by nuclear magnetic resonance techniques. The secondary structure composition can be determined via circular dichroism. Vibrational spectroscopy can also be used to characterize the conformation of peptides, polypeptides, and proteins. Cryo-electron microscopy has recently become a means of determining protein structures to high resolution, less than 5 angstroms or 0.5 nanometer, and is anticipated to increase in power as a tool for high resolution work in the next decade. This technique is still a valuable resource for researchers working with very large protein complexes such as virus coat proteins and amyloid fibers. A more qualitative picture of protein structure is often obtained by proteolysis, which is also useful to screen for more crystallizable protein samples. Novel implementations of this approach, including fast parallel proteolysis (FASTpp), can probe the structured fraction and its stability without the need for purification.
Structure classification
Protein structures can be grouped based on their similarity or a common evolutionary origin. The Structural Classification of Proteins database and CATH database provide two different structural classifications of proteins.
Computational prediction of protein structure
Main article: Protein structure predictionThe generation of a protein sequence is much easier than the determination of a protein structure. However, the structure of a protein gives much more insight in the function of the protein than its sequence. Therefore, a number of methods for the computational prediction of protein structure from its sequence have been developed. Ab initio prediction methods use just the sequence of the protein. Threading and homology modeling methods can build a 3-D model for a protein of unknown structure from experimental structures of evolutionarily-related proteins, called a protein family.
References
- Brocchieri L, Karlin S (10 June 2005). "Protein length in eukaryotic and prokaryotic proteomes". Nucleic Acids Research. 33 (10): 3390–3400. doi:10.1093/nar/gki615. PMC 1150220. PMID 15951512.
- Pauling L, Corey RB, Branson HR (1951). "The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain". Proc Natl Acad Sci USA. 37 (4): 205–211. doi:10.1073/pnas.37.4.205. PMC 1063337. PMID 14816373.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Chiang YS, Gelfand TI, Kister AE, Gelfand IM (2007). "New classification of supersecondary structures of sandwich-like proteins uncovers strict patterns of strand assemblage". Proteins. 68 (4): 915–921. doi:10.1002/prot.21473. PMID 17557333.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Govindarajan S, Recabarren R, Goldstein RA. (17 September 1999). "Estimating the total number of protein folds". Proteins. 35 (4): 408–414. doi:10.1002/(SICI)1097-0134(19990601)35:4<408::AID-PROT4>3.0.CO;2-A. PMID 10382668.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Krimm, Samuel (1986). "Vibrational Spectroscopy and Conformation of Peptides, Polypeptides, and Proteins". Advances in Protein Chemistry. Advances in Protein Chemistry. 38 (C): 181–364. doi:10.1016/S0065-3233(08)60528-8. ISBN 9780120342389.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - Minde DP, Maurice MM, Rüdiger SG (2012). Uversky, Vladimir N (ed.). "Determining biophysical protein stability in lysates by a fast proteolysis assay, FASTpp". PLoS ONE. 7 (10): e46147. doi:10.1371/journal.pone.0046147. PMC 3463568. PMID 23056252.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 7723011, please use {{cite journal}} with
|pmid=7723011instead. - Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 9309224, please use {{cite journal}} with
|pmid=9309224instead. - Zhang Y (2008). "Progress and challenges in protein structure prediction". Curr Opin Struct Biol. 18 (3): 342–348. doi:10.1016/j.sbi.2008.02.004. PMC 2680823. PMID 18436442.
Further reading
- 50 Years of Protein Structure Determination Timeline - HTML Version - National Institute of General Medical Sciences at NIH
External links
Wikis
- PDBWiki — A discussion forum for macromolecular structures (see PDBWiki)
- Proteopedia — Annotation of protein structures and other biomolecules
- TOPSAN — Annotation of protein structures in Structural genomics
Servers
- SSS Database — super-secondary structure protein database
- SPROUTS (Structural Prediction for pRotein fOlding UTility System)
- SMIR (Most Interacting Residues)
| Proteins | |
|---|---|
| Processes | |
| Structures | |
| Types | |
| Protein structural analysis | |
|---|---|
| High resolution | |
| Medium resolution | |
| Spectroscopic | |
| Translational diffusion | |
| Rotational diffusion | |
| Chemical | |
| Thermodynamic | |
| Computational | |
| ←Tertiary structureQuaternary structure→ | |
| Biomolecular structure | |
|---|---|
| Protein | |
| Nucleic acid | |
| See also | |
Template:Link GA Template:Link GA
Categories: