| 2-Succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic-acid synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 This image shows the two chains that come together to make up this enzyme This image shows the two chains that come together to make up this enzyme | |||||||||
| Identifiers | |||||||||
| EC no. | 2.2.1.9 | ||||||||
| CAS no. | 122007-88-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
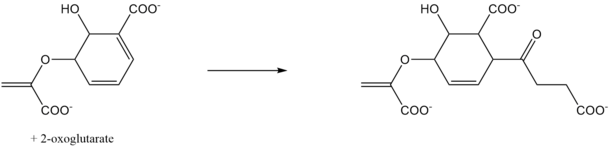
In enzymology, SEPHCHC synthase (EC EC 2.2.1.9), encoded by menD gene in E. coli, is an enzyme that catalyzes the second step of menaquinone (vitamin K2) biosynthesis. The two substrates of this enzyme are 2-oxoglutarate and isochorismate. The products of this enzyme are 5-enolpyruvoyl-6-hydroxy-2-succinyl-cyclohex-3-ene-1-carboxylate and CO2. It belongs to the transferase family.
Classification
This enzyme belongs to a family of enzymes that transfer aldehyde or ketonic groups. To be specific, this enzyme belongs to the transketolase and transaldolase families. Common names for the enzyme are:
- MenD
- SEPHCHC synthase
- 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic-acid synthase
- Systematic name: isochorismate:2-oxoglutarate 4-oxopentanoatetransferase
Reaction
In the biosynthesis of vitamin K, SEPHCHC is involved in the second step of the pathway. The type of reaction is decarboxylating, and to have maximum activity, this enzymes uses the cofactor Mg2+, a magnesium ion. In previous years, it was thought that this reaction led to SHCHC, the product MenH. After further research, we now know that this reaction is a new step in the pathway. The actual product of this enzyme can lose a pyruvate spontaneously.
Structure
The structures reveal a stable dimer-of-dimers association in agreement with gel filtration and analytical studies confirm the classification of MenD in the pyruvate oxidase family. The active site is highly basic with a hydrophobic patch. These features match with the chemical properties of the substrates.
Homologues
There are many similar structures MenD. Though it is commonly found in E. Coli, but can be found in other organisms as well. Bacillus subtilis and Mycobacterium tuberculosis are two homologues. All of the organisms share something in common, being that catalyze decarboxylation reactions.
References
- "Enzyme entry EC: 2.2.1.9". ExPASy.
- "The Enzyme List Class 2" (PDF). September 2010. Retrieved 25 August 2016.
- "Enzyme 2.2.1.9". KEGG.
- PDB: 2jla; Dawson A, Fyfe PK, Hunter WN (December 2008). "Specificity and reactivity in menaquinone biosynthesis: the structure of Escherichia coli MenD (2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase)". Journal of Molecular Biology. 384 (5): 1353–68. doi:10.1016/j.jmb.2008.10.048. PMC 3656419. PMID 18983854.
| Transferases: aldehyde-ketone (EC 2.2) | |
|---|---|
| 2.2.1 | |
| Enzymes | |
|---|---|
| Activity | |
| Regulation | |
| Classification | |
| Kinetics | |
| Types |
|