 | |
| Identifiers | |
|---|---|
IUPAC name
| |
| CAS Number | |
| PubChem CID | |
| IUPHAR/BPS | |
| DrugBank | |
| ChemSpider | |
| UNII | |
| CompTox Dashboard (EPA) | |
| Chemical and physical data | |
| Formula | C28H26N2O10 |
| Molar mass | 550.520 g·mol |
| (what is this?) (verify) | |
Balanol is a fungal metabolite produced by the fungus Verticillium balanoides. It is a potent inhibitor of the serine/threonine kinases protein kinase A (PKA) and protein kinase C (PKC), binding in a similar manner with that of ATP. Balanol was discovered in 1993 in the search for novel inhibitors of PKC, a member of a family of serine/threonine kinases whose overactivation is associated with numerous human diseases of signal transduction including cancer. However, much of the research on balanol focuses on how chemical modifications of the molecular structure affect binding to PKA. Indeed, balanol, its chemically altered analogs, and their interactions with PKA in particular are used to illuminate the roles of selectivity and protein flexibility in the inhibition of kinases. For instance, the X-ray crystal structure of balanol in complex with PKA was used in order to confer selectivity and to improve pharmacological efficacy of inhibitors of the H. sapiens Akt (PKB), another serine/threonine protein kinase implicated in the proper functioning of many cellular processes.
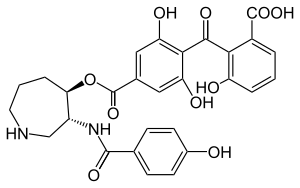
Structure
The chemical structure was initially characterized through a combination of IR spectroscopy, hydrogen-1 NMR, carbon-13 NMR, and 2D NMR spectra data, and the crystal structure of balanol in complex with PKA was solved in 1999. The balanol molecule consists of three regions: a benzophenone, hexahydroazepane, and 4-hydroxy benzoyl moiety. The benzophenone and hexahydroazepane moieties are connected via an ester linkage, and the azepane and benzoyl moieties are connected through an amide linkage. Furthermore, balanol is sometimes referred to as ophiocordin, an antifungal agent produced by the fungus Cordyceps ophioglossoides whose structure is regioisometric to that of balanol; that is, in ophiocordin, the benzophenone is linked to the hexahydroazepane instead via an amide; the 4-hydroxy benzoyl group and hexahydroazepane are connected with an ester linkage.
Balanol is a congener of ATP, with distinct regions of its molecular structure able to form bonds like the adenine ring, ribose, and phosphate groups of the ATP molecule. Specifically, balanol's 4-hydroxybenzamide moiety including the amide linker corresponds to the adenine of ATP, the hexahydroazepane moiety to the ribose region, and the benzophenone rings to ATP's triphosphates.
Numerous balanol congeners have been synthesized in order to study the effect of chemical modifications on kinase binding and specificity. For example, modifications of the benzophenone rings, analogs of the phosphate groups of ATP, produce unusually potent and specific protein kinase inhibitors. Additionally, many of these balanol congeners show substantial specificity toward PKA over PKC. Elimination of the hydroxyl group from the benzophenone ring (producing 10"-deoxybalanol), for instance, elicits selectivity two orders of magnitude greater for PKA over PKC. Potent inhibitory activity was also shown to be dependent on the carboxylic acid of the benzophenone ring and the presence of a five- or seven-membered azepane ring.
Balanol's flexible structure also plays an important role in its selectivity. Specifically, balanol's distal benzophenone ring is able to rotate. In fact, in complex with PKA, balanol's distal benzophenone ring was observed to be nearly orthogonal with the neighboring ring. This flexibility could allow balanol to adapt to numerous protein microenvironments in order to exert its inhibitory properties on various protein kinases. Balanol's selectivity for some kinases and not others, in turn, could represent the varying degrees of flexibilities allowed by these kinases' catalytic ATP-binding sites.
Biological activities
Balanol is one of the most potent, naturally occurring inhibitors of protein kinases PKC and PKA. Balanol was originally discovered to inhibit PKC and many of its isoforms in humans (α, β-Ι, β-ΙΙ, γ, δ, ε, η), with an inhibition profile similar to that of staurosporine.
Balanol prevents functioning by binding competitively to the catalytic domain of PKC and PKA with an affinity (Ki ≥ 4 nM) three orders of magnitude greater than for ATP. Without ATP bound, these kinases are unable to catalyze the transfer of the γ-phosphate from ATP to the kinase's target substrate, and thus function is hindered.
Binding
Bound balanol extends 17.2 Å from the hydroxyl oxygen atom of its 4-hydroxybenzamide to the most distal carboxyl oxygen atom of its benzophenone moiety, fitting between the large and small catalytic lobes of PKA and extending from the inner edge to the outer mouth of the active-site ATP-binding cleft of the enzyme. The cleft's glycine-rich loop and small lobe allow for tight, induced-fit binding.
Each subsite of the ATP-binding pocket in PKA is occupied by a substituent of the balanol molecule as observed in studies of recombinant mouse catalytic subunit of PKA. The adenine subsite of PKA has hydrophobic components and has the potential to donate and accept electrons to make hydrogen bonds with one or two planar cyclic rings. Specifically, the backbone nitrogen of Val123 and the carbonyl oxygen atom of Glu121 can form hydrogen bonds with balanol's single hydroxyl group of the 4-hydroxyl benzoyl moiety. In the ATP molecule, the Val123 amide hydrogen atom accepts electrons from ATP's purine ring N1 atom while the Glu121 backbone carbonyl oxygen atom donates electrons to the ATP purine ring N6 atom. Mimetics of ATP like balanol can be anchored within the hydrophobic adenine-binding pocket because their planar substituents can make favorable nonpolar interactions. The ribose subsite is occupied by balanol's axepane ring. Specifically, the N1 atom of the azepane ring hydrogen bonds with the backbone carbonyl oxygen atom of Glu170 of the catalytic loop. Atoms within the azepane ring also can make favorable nonpolar contacts with residues Gly50, Flu127, and Glu170. While other interactions also occur, most of the polar and nonpolar interactions involve the benzophenone substituent of balanol, which interacts with residues comprising the phosphate-binding subsite. For instance, balanol's benzophenone ring interacts with numerous highly conserved residues in PKA such as Gly52, Phe54, Asp184 among others in addition to various nonconserved residues such as Leu74 and Gln84. Most of the conserved residues in PKA interact with ATP whereas interactions of ATP with the nonconserved residues do not occur.
Six ordered water molecules also play critical roles in balanol binding to PKA. Several PKA residues such as Leu49 and Tyr330 hydrogen bond with conserved water molecules. These water molecules also mediate interactions between balanol and the residues of the catalytic cleft of PKA; for example, two water molecules form a bridge to allow balanol's N1’ atom to interact with the hydroxyl group of Tyr330.
However, the tight interaction of balanol with the ATP-binding site that leads to balanol's potency as an inhibitor is not due to hydrogen bonding but rather because of the predominant nonpolar interactions the molecule makes. For instance, balanol's benzophenone rings are positioned beside the glycine-rich loop and further from the more polar catalytic loop. Compared to ATP's triphosphates, accommodating balanol's benzophenone rings induces the rearrangement of catalytic side-chains Phe54 and Ser53, enabling favorable polar and nonpolar interactions and likely contributing to balanol's potent inhibitory effect on these kinases.
Action against other protein kinases
As balanol acts specifically at the ATP-binding site in the catalytic core of PKA and PKC, it was initially hypothesized that balanol could inhibit all serine/threonine kinases sharing a conserved ATP-binding site. However, balanol does not inhibit all serine/threonine kinases uniformly. Affinity to balanol varies greatly (Ki = 1.6–742 nM), with several members of the serine/threonine protein kinase family not being affected at all, even though affinity of these kinases to ATP varies little (13–60 μM). Balanol exhibits the most potent inhibitory effect (Ki = 1.6–6.4 nM) on cGMP-dependent protein kinase (PKG), PKA, and PKC, including isoforms. It exerts much less of an effect (Ki = 30 – 742 nM), if any, on Ca-calmodulin-regulated kinases, mitogen-activating protein kinase (MAPK/Erk1), and certain cyclin-dependent kinases. Furthermore, balanol does not inhibit two tyrosine protein kinases, the SC kinase, nor the epidermal growth factor receptor kinase.
References
- ^ Kulanthaivel P, Hallock YF, Boros C, Hamilton SM, Janzen WP, Ballas LM, Loomis CR, Jiang JB, Katz B (1993). "Balanol: A novel and potent inhibitor of protein kinase C from the fungus Verticillium balanoides". J. Am. Chem. Soc. 115 (14): 6452–6453. doi:10.1021/ja00067a087.
- ^ Koide K, Bunnage ME, Gomez Paloma L, Kanter JR, Taylor SS, Brunton LL, Nicolaou KC (September 1995). "Molecular design and biological activity of potent and selective protein kinase inhibitors related to balanol". Chemistry & Biology. 2 (9): 601–8. doi:10.1016/1074-5521(95)90124-8. PMID 9383464.
- ^ Pande V, Ramos MJ, Gago F (August 2008). "The Protein kinase inhibitor balanol: structure-activity relationships and structure-based computational studies". Anti-Cancer Agents in Medicinal Chemistry. 8 (6): 638–45. doi:10.2174/187152008785133056. PMID 18690827.
- Gassel M, Breitenlechner CB, Rüger P, Jucknischke U, Schneider T, Huber R, et al. (June 2003). "Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)". Journal of Molecular Biology. 329 (5): 1021–34. doi:10.1016/S0022-2836(03)00518-7. PMID 12798691.
- ^ Narayana N, Diller TC, Koide K, Bunnage ME, Nicolaou KC, Brunton LL, et al. (February 1999). "Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase". Biochemistry. 38 (8): 2367–76. doi:10.1021/bi9820659. PMID 10029530.
- ^ Setyawan J, Koide K, Diller TC, Bunnage ME, Taylor SS, Nicolaou KC, Brunton LL (August 1999). "Inhibition of protein kinases by balanol: specificity within the serine/threonine protein kinase subfamily". Molecular Pharmacology. 56 (2): 370–6. doi:10.1124/mol.56.2.370. PMID 10419556.
- Saha T, Maitra R, Chattopadhyay SK (December 2013). "A unified approach to the important protein kinase inhibitor balanol and a proposed analogue". Beilstein Journal of Organic Chemistry. 9: 2910–5. doi:10.3762/bjoc.9.327. PMC 3896276. PMID 24454570.
- Gustafsson AB, Brunton LL (August 1999). "Differential and selective inhibition of protein kinase A and protein kinase C in intact cells by balanol congeners". Molecular Pharmacology. 56 (2): 377–82. doi:10.1124/mol.56.2.377. PMID 10419557.