Cell-free fetal DNA (cffDNA) is fetal DNA that circulates freely in the maternal blood. Maternal blood is sampled by venipuncture. Analysis of cffDNA is a method of non-invasive prenatal diagnosis frequently ordered for pregnant women of advanced maternal age. Two hours after delivery, cffDNA is no longer detectable in maternal blood.
Background
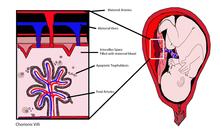
cffDNA originates from placental trophoblasts. Fetal DNA is fragmented when placental microparticles are shed into the maternal blood circulation.
cffDNA fragments are approximately 200 base pairs (bp) in length. They are significantly smaller than maternal DNA fragments. The difference in size allows cffDNA to be distinguished from maternal DNA fragments.
Approximately 11 to 13.4 percent of the cell-free DNA in maternal blood is of fetal origin. The amount varies widely from one pregnant woman to another. cffDNA is present after five to seven weeks gestation. The amount of cffDNA increases as the pregnancy progresses. The quantity of cffDNA in maternal blood diminishes rapidly after childbirth. Two hours after delivery, cffDNA is no longer detectable in maternal blood.
Analysis of cffDNA may provide earlier diagnosis of fetal conditions than current techniques. As cffDNA is found in maternal blood, sampling carries no associated risk of spontaneous abortion. cffDNA analysis has the same ethical and practical issues as other techniques such as amniocentesis and chorionic villus sampling.
Some disadvantages of sampling cffDNA include a low concentration of cffDNA in maternal blood; variation in the quantity of cffDNA between individuals; a high concentration of maternal cell free DNA compared to the cffDNA in maternal blood.
New evidence shows that cffDNA test failure rate is higher, fetal fraction (proportion of fetal versus maternal DNA in the maternal blood sample) is lower and PPV for trisomies 18, 13 and SCA is decreased in IVF pregnancies compared to those conceived spontaneously.
Laboratory methods
A number of laboratory methods have been developed for cell-free fetal DNA screening for genetic defects have been developed. The main ones are (1) massively parallel shotgun sequencing (MPSS), (2) targeted massive parallel sequencing (t-MPS) and (3) single nucleotide polymorphism (SNP) based approach.
A maternal peripheral blood sample is taken by venesection at about ten weeks gestation.
Separation of cffDNA
Blood plasma is separated from the maternal blood sample using a laboratory centrifuge. The cffDNA is then isolated and purified. A standardized protocol for doing this was written through an evaluation of the scientific literature. The highest yield in cffDNA extraction was obtained with the "QIAamp DSP Virus Kit".
Addition of formaldehyde to maternal blood samples increases the yield of cffDNA. Formaldehyde stabilizes intact cells, and therefore inhibits the further release of maternal DNA. With the addition of formaldehyde, the percentage of cffDNA recovered from a maternal blood sample varies between 0.32 percent and 40 percent with a mean of 7.7 percent. Without the addition of formaldehyde, the mean percentage of cffDNA recovered has been measured at 20.2 percent. However, other figures vary between 5 and 96 percent.
Recovery of cffDNA may be related to the length of the DNA fragments. Another way to increase the fetal DNA is based on physical length of DNA fragments. Smaller fragments can represent up to seventy percent of the total cell free DNA in the maternal blood sample.
Analysis of cffDNA
In real-time PCR, fluorescent probes are used to monitor the accumulation of amplicons. The reporter fluorescent signal is proportional to the number of amplicons generated. The most appropriate real time PCR protocol is designed according to the particular mutation or genotype to be detected. Point mutations are analysed with qualitative real time PCR with the use of allele specific probes. insertions and deletions are analyzed by dosage measurements using quantitative real time PCR.
cffDNA may be detected by finding paternally inherited DNA sequences via polymerase chain reaction (PCR).
Quantitative real-time PCR
sex-determining region Y gene (SRY) and Y chromosome short tandem repeat "DYS14" in cffDNA from 511 pregnancies were analyzed using quantitative real-time PCR (RT-qPCR). In 401 of 403 pregnancies where maternal blood was drawn at seven weeks gestation or more, both segments of DNA were found.
Nested PCR
The use of nested polymerase chain reaction (nested PCR) was evaluated to determine sex by detecting a Y chromosome specific signal in the cffDNA from maternal plasma. Nested PCR detected 53 of 55 male fetuses. The cffDNA from the plasma of 3 of 25 women with female fetuses contained the Y chromosome-specific signal. The sensitivity of nested PCR in this experiment was 96 percent. The specificity was 88 percent.
Digital PCR
Microfluidic devices allow the quantification of cffDNA segments in maternal plasma with accuracy beyond that of real-time PCR. Point mutations, loss of heterozygosity and aneuploidy can be detected in a single PCR step. Digital PCR can differentiate between maternal blood plasma and fetal DNA in a multiplex fashion.
Shotgun sequencing
High throughput shotgun sequencing using tools such as Solexa or Illumina, yields approximately 5 million sequence tags per sample of maternal serum. Aneuploid pregnancies such as trisomy were identified when testing at the fourteenth week of gestation. Fetal whole of genome mapping by parental haplotype analysis was completed using sequencing of cffDNA from maternal serum. Pregnant females were studied using a 2-plex massively parallel maternal plasma DNA sequencing and trisomy was diagnosed with z-score greater than 3. The sequencing gave sensitivity of 100 percent, specificity of 97.9 percent, a positive predictive value of 96.6 percent and a negative predictive value of 100 percent.
Mass spectrometry
Matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) combined with single-base extension after PCR allows cffDNA detection with single base specificity and single DNA molecule sensitivity. DNA is amplified by PCR. Then, linear amplification with base extension reaction (with a third primer) is designed to anneal to the region upstream from the mutation site. One or two bases are added to the extension primer to produce two extension products from wild-type DNA and mutant DNA. Single base specificity provides advantages over hybridization-based techniques using TaqMan hydrolysis probes. When assessing the technique, no false positives or negatives were found when looking for cffDNA to determine fetal sex in sixteen maternal plasma samples. The sex of ninety-one male foetuses were correctly detected using MALDI-TOF mass spectrometry. The technique had accuracy, sensitivity and specificity of over 99 percent.
Epigenetic modifications
Differences in gene activation between maternal and fetal DNA can be exploited. Epigenetic modifications (heritable modifications that change gene function without changing DNA sequence) can be used to detect cffDNA. The hypermethylated RASSF1A promoter is a universal fetal marker used to confirm the presence of cffDNA. A technique was described where cffDNA was extracted from maternal plasma and then digested with methylation-sensitive and insensitive restriction enzymes. Then, real-time PCR analysis of RASSF1A, SRY, and DYS14 was done. The procedure detected 79 out of 90 (88 percent) maternal blood samples where hypermethylated RASSF1A was present.
mRNA
mRNA transcripts from genes expressed in the placenta are detectable in maternal plasma. In this procedure, plasma is centrifuged so an aqueous layer appears. This layer is transferred and from it RNA is extracted. RT-PCR is used to detect a selected expression of RNA. For example, Human placental lactogen (hPL) and beta-hCG mRNA are stable in maternal plasma and can be detected. (Ng et al. 2002). This can help to confirm the presence of cffDNA in maternal plasma.
Applications
Prenatal sex discernment
Further information: Prenatal sex discernmentThe analysis of cffDNA from a sample of maternal plasma allows for prenatal sex discernment. Applications of prenatal sex discernment include:
- Disease testing: Whether the sex of the fetus is male or female allows the determination of the risk of a particular X-linked recessive genetic disorder in a particular pregnancy, especially where the mother is a genetic carrier of the disorder.
- Preparation, for any sex-dependent aspects of parenting.
- Sex selection, which after preimplantation genetic diagnosis may be performed by selecting only embryos of the preferred sex, or, after post-implantation methods by performing sex-selective abortion depending on the test result and personal preference.
In comparison to obstetric ultrasonography which is unreliable for sex determination in the first trimester and amniocentesis which carries a small risk of miscarriage, sampling of maternal plasma for analysis of cffDNA is without risk. The main targets in the cffDNA analysis are the gene responsible for the sex-determining region Y protein (SRY) on the Y chromosome and the DYS14 sequence.
Congenital adrenal hyperplasia
In congenital adrenal hyperplasia, the adrenal cortex lacks appropriate corticosteroid synthesis, leading to excess adrenal androgens and affects female fetuses. There is an external masculinization of the genitalia in the female fetuses. Mothers of at risk fetuses are given dexamethasone at 6 weeks gestation to suppress pituitary gland release of androgens.
If analysis of cffDNA obtained from a sample of maternal plasma lacks genetic markers found only on the Y chromosome, it is suggestive of a female fetus. However, it might also indicate a failure of the analysis itself (a false negative result). Paternal genetic polymorphisms and sex-independent markers may be used to detect cffDNA. A high degree of heterozygosity of these markers must be present for this application.
Paternity testing
Prenatal DNA paternity testing is commercially available. The test can be performed at nine weeks gestation.
Single gene disorders
Autosomal dominant and recessive single gene disorders which have been diagnosed prenatally by analysing paternally inherited DNA include cystic fibrosis, beta thalassemia, sickle cell anemia, spinal muscular atrophy, and myotonic dystrophy. Prenatal diagnosis of single gene disorders which are due to an autosomal recessive mutation, a maternally inherited autosomal dominant mutation or large sequence mutations that include duplication, expansion or insertion of DNA sequences is more difficult.
In cffDNA, fragments of 200 – 300 bp length involved in single gene disorders are more difficult to detect.
For example, the autosomal dominant condition, achondroplasia is caused by the FGFR3 gene point mutation. In two pregnancies with a fetus with achondroplasia was found a paternally inherited G1138A mutation from cffDNA from a maternal plasma sample in one and a G1138A de novo mutation from the other.
In studies of the genetics of Huntington's chorea using qRT-PCR of cffDNA from maternal plasma samples, CAG repeats have been detected at normal levels (17, 20 and 24).
cffDNA may also be used to diagnose single gene disorders. Developments in laboratory processes using cffDNA may allow prenatal diagnosis of aneuploidies such as trisomy 21 (Down's syndrome) in the fetus.
Hemolytic disease of the fetus and newborn
Incompatibility of fetal and maternal RhD antigens is the main cause of Hemolytic disease of the newborn. Approximately 15 percent of Caucasian women, 3 to 5 percent of black Africa women and less than 3 percent of Asian women are RhD negative.
Accurate prenatal diagnosis is important because the disease can be fatal to the newborn and because treatment including intramuscular immunoglobulin (Anti-D) or intravenous immunoglobulin can be administered to mothers at risk.
PCR to detect RHD (gene) gene exons 5 and 7 from cffDNA obtained from maternal plasma between 9 and 13 weeks gestation gives a high degree of specificity, sensitivity and diagnostic accuracy (>90 percent) when compared to RhD determination from newborn cord blood serum. Similar results were obtained targeting exons 7 and 10. Droplet digital PCR in fetal RhD determination was comparable to a routine real-time PCR technique.
Routine determination of fetal RhD status from cffDNA in maternal serum allows early management of at risk pregnancies while decreasing unnecessary use of Anti-D by over 25 percent.
Aneuploidy
- Sex chromosomes
Analysis of maternal serum cffDNA by high-throughput sequencing can detect common fetal sex chromosome aneuploidies such as Turner's syndrome, Klinefelter's syndrome and triple X syndrome but the procedure's positive predictive value is low.
- Trisomy 21
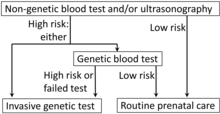
Fetal trisomy of chromosome 21 is the cause of Down's syndrome. This trisomy can be detected by analysis of cffDNA from maternal blood by massively parallel shotgun sequencing (MPSS). Another technique is digital analysis of selected regions (DANSR). Such tests show a sensitivity of about 99% and a specificity of more than 99.9%. Therefore, they cannot be regarded as diagnostic procedures but may be used to confirm a positive maternal screening test such as a first trimester screening or ultrasound markers of the condition.
- Trisomy 13 and 18
Analysis of cffDNA from maternal plasma with MPSS looking for trisomy 13 or 18 is possible
Factors limiting sensitivity and specificity include the levels of cffDNA in the maternal plasma; maternal chromosomes may have mosaicism.
A number of fetal nucleic acid molecules derived from aneuploid chromosomes can be detected including SERPINEB2 mRNA, clad B, hypomethylated SERPINB5 from chromosome 18, placenta-specific 4 (PLAC4), hypermethylated holocarboxylase synthetase (HLCS) and c21orf105 mRNA from chromosome 12. With complete trisomy, the mRNA alleles in maternal plasma isn't the normal 1:1 ratio, but is in fact 2:1. Allelic ratios determined by epigenetic markers can also be used to detect the complete trisomies. Massive parallel sequencing and digital PCR for fetal aneuploidy detection can be used without restriction to fetal-specific nucleic acid molecules. (MPSS) is estimated to have a sensitivity of between 96 and 100%, and a specificity between 94 and 100% for detecting Down syndrome. It can be performed at 10 weeks of gestational age. One study in the United States estimated a false positive rate of 0.3% and a positive predictive value of 80% when using cffDNA to detect Down syndrome.
Preeclampsia
Preeclampsia is a complex condition of pregnancy involving hypertension and proteinuria usually after 20 weeks gestation. It is associated with poor cytotrophoblastic invasion of the myometrium. Onset of the condition between 20 and 34 weeks gestation, is considered "early". Maternal plasma samples in pregnancies complicated by preeclampsia have significantly higher levels of cffDNA that those in normal pregnancies. This holds true for early onset preeclampsia.
History
In 1997, Hong Kong molecular biologist Dennis Lo and his team first employed the Y-PCR assay to identify fetal Y chromosome sequences (because Y-specific sequences are genetic sequences of the fetus not in the maternal genome) in maternal plasma samples. For this groundbreaking work, Lo was honored with the 2022 Lasker DeBakey Clinical Medical Research Award.
Future perspectives
New generation sequencing may be used to yield a whole genome sequence from cffDNA. This raises ethical questions. However, the utility of the procedure may increase as clear associations between specific genetic variants and disease states are discovered.
See also
References
- Alberry M, Maddocks D, Jones M, Abdel Hadi M, Abdel-Fattah S, Avent N, Soothill PW (May 2007). "Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast". Prenatal Diagnosis. 27 (5). Wiley-Blackwell: 415–8. doi:10.1002/pd.1700. PMID 17286310. S2CID 39693586.
- Gupta AK, Holzgreve W, Huppertz B, Malek A, Schneider H, Hahn S (November 2004). "Detection of fetal DNA and RNA in placenta-derived syncytiotrophoblast microparticles generated in vitro". Clinical Chemistry. 50 (11). American Association for Clinical Chemistry (AACC): 2187–90. doi:10.1373/clinchem.2004.040196. PMID 15502097.
- Smets EM, Visser A, Go AT, van Vugt JM, Oudejans CB (February 2006). "Novel biomarkers in preeclampsia". Clinica Chimica Acta; International Journal of Clinical Chemistry. 364 (1–2). Elsevier BV: 22–32. doi:10.1016/j.cca.2005.06.011. PMID 16139262.
- Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM (January 2004). "Size distributions of maternal and fetal DNA in maternal plasma". Clinical Chemistry. 50 (1). American Association for Clinical Chemistry (AACC): 88–92. doi:10.1373/clinchem.2003.024893. PMID 14709639.
- Li Y, Zimmermann B, Rusterholz C, Kang A, Holzgreve W, Hahn S (June 2004). "Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms" (PDF). Clinical Chemistry. 50 (6). American Association for Clinical Chemistry (AACC): 1002–11. doi:10.1373/clinchem.2003.029835. PMID 15073090.
- Li Y, Di Naro E, Vitucci A, Zimmermann B, Holzgreve W, Hahn S (February 2005). "Detection of paternally inherited fetal point mutations for beta-thalassemia using size-fractionated cell-free DNA in maternal plasma". JAMA. 293 (7). American Medical Association (AMA): 843–9. doi:10.1001/jama.293.7.843. PMID 15713774.
- Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A (July 2013). "Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma". Prenatal Diagnosis. 33 (7): 662–6. doi:10.1002/pd.4119. PMID 23553731. S2CID 31630351.
- Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM (April 1998). "Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis". American Journal of Human Genetics. 62 (4). Elsevier BV: 768–75. doi:10.1086/301800. PMC 1377040. PMID 9529358.
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM (January 1999). "Rapid clearance of fetal DNA from maternal plasma". American Journal of Human Genetics. 64 (1). Elsevier BV: 218–24. doi:10.1086/302205. PMC 1377720. PMID 9915961.
- Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, Poon PM, Redman CW, Wainscoat JS (December 1998). "Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma". The New England Journal of Medicine. 339 (24). New England Journal of Medicine (NEJM/MMS): 1734–8. doi:10.1056/nejm199812103392402. PMID 9845707.
- Allyse M, Sayres LC, King JS, Norton ME, Cho MK (November 2012). "Cell-free fetal DNA testing for fetal aneuploidy and beyond: clinical integration challenges in the US context". Human Reproduction. 27 (11). Oxford University Press (OUP): 3123–31. doi:10.1093/humrep/des286. PMC 3472618. PMID 22863603.
- Mujezinovic F, Alfirevic Z (September 2007). "Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review". Obstetrics and Gynecology. 110 (3). Ovid Technologies (Wolters Kluwer Health): 687–94. doi:10.1097/01.aog.0000278820.54029.e3. PMID 17766619. S2CID 25548568.
- ^ Lo YM (August 2008). "Fetal nucleic acids in maternal plasma". Annals of the New York Academy of Sciences. 1137 (1). Wiley-Blackwell: 140–3. Bibcode:2008NYASA1137..140L. doi:10.1196/annals.1448.004. PMID 18837938. S2CID 3445205.
- "Reliable Accurate Prenatal non-Invasive Diagnosis". The NHS RAPID project. Archived from the original on 2019-03-01. Retrieved 2016-07-08.
- ^ Hahn S, Chitty LS (April 2008). "Noninvasive prenatal diagnosis: current practice and future perspectives". Current Opinion in Obstetrics & Gynecology. 20 (2): 146–51. doi:10.1097/GCO.0b013e3282f73349. PMID 18388814. S2CID 7222299.
- ^ Wright CF, Burton H (22 October 2008). "The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis". Human Reproduction Update. 15 (1). Oxford University Press (OUP): 139–51. doi:10.1093/humupd/dmn047. PMID 18945714.
- Lee TJ, Rolnik DL, Menezes MA, McLennan AC, da Silva Costa F (April 2018). "Cell-free fetal DNA testing in singleton IVF conceptions". Human Reproduction. 33 (4): 572–578. doi:10.1093/humrep/dey033. PMID 29462319.
- Dar P, Shani H, Evans MI (June 2016). "Cell-free DNA: Comparison of Technologies". Clinics in Laboratory Medicine. 36 (2): 199–211. doi:10.1016/j.cll.2016.01.015. PMID 27235906.
- Grace MR, Hardisty E, Dotters-Katz SK, Vora NL, Kuller JA (August 2016). "Cell-Free DNA Screening: Complexities and Challenges of Clinical Implementation". Obstetrical & Gynecological Survey. 71 (8): 477–87. doi:10.1097/OGX.0000000000000342. PMC 5548289. PMID 27526871.
- Allen S, Young E, Bowns B (April 2017). "Noninvasive prenatal diagnosis for single gene disorders". Current Opinion in Obstetrics & Gynecology. 29 (2): 73–79. doi:10.1097/GCO.0000000000000347. PMID 28134670. S2CID 33474139.
- Guibert J, Benachi A, Grebille AG, Ernault P, Zorn JR, Costa JM (August 2003). "Kinetics of SRY gene appearance in maternal serum: detection by real time PCR in early pregnancy after assisted reproductive technique". Human Reproduction. 18 (8): 1733–6. doi:10.1093/humrep/deg320. PMID 12871892.
- Chiu RW, Poon LL, Lau TK, Leung TN, Wong EM, Lo YM (September 2001). "Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma". Clinical Chemistry. 47 (9): 1607–13. doi:10.1093/clinchem/47.9.1607. PMID 11514393.
- Legler TJ, Liu Z, Mavrou A, Finning K, Hromadnikova I, Galbiati S, Meaney C, Hultén MA, Crea F, Olsson ML, Maddocks DG, Huang D, Fisher SA, Sprenger-Haussels M, Soussan AA, van der Schoot CE (September 2007). "Workshop report on the extraction of foetal DNA from maternal plasma". Prenatal Diagnosis. 27 (9). Wiley-Blackwell: 824–9. doi:10.1002/pd.1783. PMID 17604339. S2CID 38860225.
- Dhallan R, Au WC, Mattagajasingh S, Emche S, Bayliss P, Damewood M, Cronin M, Chou V, Mohr M (March 2004). "Methods to increase the percentage of free fetal DNA recovered from the maternal circulation". JAMA. 291 (9). American Medical Association (AMA): 1114–9. doi:10.1001/jama.291.9.1114. PMID 14996781.
- Benachi A, Yamgnane A, Olivi M, Dumez Y, Gautier E, Costa JM (January 2005). "Impact of formaldehyde on the in vitro proportion of fetal DNA in maternal plasma and serum". Clinical Chemistry. 51 (1). American Association for Clinical Chemistry (AACC): 242–4. doi:10.1373/clinchem.2004.038125. PMID 15514098.
- Chinnapapagari SK, Holzgreve W, Lapaire O, Zimmermann B, Hahn S (March 2005). "Treatment of maternal blood samples with formaldehyde does not alter the proportion of circulatory fetal nucleic acids (DNA and mRNA) in maternal plasma". Clinical Chemistry. 51 (3). American Association for Clinical Chemistry (AACC): 652–5. doi:10.1373/clinchem.2004.042119. PMID 15738521.
- ^ Traeger-Synodinos J (2006). "Real-time PCR for prenatal and preimplantation genetic diagnosis of monogenic diseases". Molecular Aspects of Medicine. 27 (2–3). Elsevier BV: 176–91. doi:10.1016/j.mam.2005.12.004. PMID 16430951.
- Boon EM, Schlecht HB, Martin P, Daniels G, Vossen RH, den Dunnen JT, Bakker B, Elles R (October 2007). "Y chromosome detection by Real Time PCR and pyrophosphorolysis-activated polymerisation using free fetal DNA isolated from maternal plasma". Prenatal Diagnosis. 27 (10). Wiley-Blackwell: 932–7. doi:10.1002/pd.1804. PMID 17600849. S2CID 24498216.
- Hill M, Pařízek A, Cibula D, Kancheva R, Jirásek JE, Jirkovská M, Velíková M, Kubátová J, Klímková M, Pašková A, Zižka Z, Kancheva L, Kazihnitková H, Zamrazilová L, Stárka L (October 2010). "Steroid metabolome in fetal and maternal body fluids in human late pregnancy". The Journal of Steroid Biochemistry and Molecular Biology. 122 (4). Elsevier BV: 114–32. doi:10.1016/j.jsbmb.2010.05.007. PMID 20580824. S2CID 25820012.
- Al-Yatama MK, Mustafa AS, Ali S, Abraham S, Khan Z, Khaja N (May 2001). "Detection of Y chromosome-specific DNA in the plasma and urine of pregnant women using nested polymerase chain reaction". Prenatal Diagnosis. 21 (5). Wiley-Blackwell: 399–402. doi:10.1002/pd.69. PMID 11360283. S2CID 20169086.
- ^ Zimmermann BG, Grill S, Holzgreve W, Zhong XY, Jackson LG, Hahn S (December 2008). "Digital PCR: a powerful new tool for noninvasive prenatal diagnosis?". Prenatal Diagnosis. 28 (12). Wiley-Blackwell: 1087–93. doi:10.1002/pd.2150. PMID 19003785. S2CID 2909830.
- ^ Lo YM, Lun FM, Chan KC, Tsui NB, Chong KC, Lau TK, Leung TY, Zee BC, Cantor CR, Chiu RW (August 2007). "Digital PCR for the molecular detection of fetal chromosomal aneuploidy". Proceedings of the National Academy of Sciences of the United States of America. 104 (32). Proceedings of the National Academy of Sciences: 13116–21. Bibcode:2007PNAS..10413116L. doi:10.1073/pnas.0705765104. PMC 1934923. PMID 17664418.
- Quake S (July 2007). "At the interface of physics and biology". BioTechniques. 43 (1): 19. PMID 17695250.
- Chiu RW, Lo YM (November 2010). "Pregnancy-associated microRNAs in maternal plasma: a channel for fetal-maternal communication?". Clinical Chemistry. 56 (11). American Association for Clinical Chemistry (AACC): 1656–7. doi:10.1373/clinchem.2010.153684. PMID 20837782.
- ^ Ding C (2008). "MALDI-TOF Mass Spectrometry for Analyzing Cell-Free Fetal DNA in Maternal Plasma". Prenatal Diagnosis. Methods in Molecular Biology. Vol. 444. Totowa, NJ: Humana Press. pp. 253–67. doi:10.1007/978-1-59745-066-9_20. ISBN 978-1-58829-803-4. PMID 18425487.
- Akolekar R, Farkas DH, VanAgtmael AL, Bombard AT, Nicolaides KH (October 2010). "Fetal sex determination using circulating cell-free fetal DNA (ccffDNA) at 11 to 13 weeks of gestation". Prenatal Diagnosis. 30 (10). Wiley-Blackwell: 918–23. doi:10.1002/pd.2582. PMID 20721878. S2CID 20744999.
- Tong YK, Chiu RW, Chan KC, Leung TY, Lo YM (September 2012). "Technical concerns about immunoprecipitation of methylated fetal DNA for noninvasive trisomy 21 diagnosis". Nature Medicine. 18 (9). Springer Nature: 1327–8, author reply 1328–9. doi:10.1038/nm.2915. PMID 22961155. S2CID 31316176.
- Papageorgiou EA, Karagrigoriou A, Tsaliki E, Velissariou V, Carter NP, Patsalis PC (April 2011). "Fetal-specific DNA methylation ratio permits noninvasive prenatal diagnosis of trisomy 21". Nature Medicine. 17 (4). Springer Nature: 510–3. doi:10.1038/nm.2312. PMC 3977039. PMID 21378977.
- ^ White HE, Dent CL, Hall VJ, Crolla JA, Chitty LS (14 September 2012). Oudejans C (ed.). "Evaluation of a novel assay for detection of the fetal marker RASSF1A: facilitating improved diagnostic reliability of noninvasive prenatal diagnosis". PLOS ONE. 7 (9). Public Library of Science (PLoS): e45073. Bibcode:2012PLoSO...745073W. doi:10.1371/journal.pone.0045073. PMC 3443218. PMID 23024794.
- Ng EK, Tsui NB, Lam NY, Chiu RW, Yu SC, Wong SC, Lo ES, Rainer TH, Johnson PJ, Lo YM (August 2002). "Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals". Clinical Chemistry. 48 (8): 1212–7. doi:10.1093/clinchem/48.8.1212. PMID 12142376.
- Baird PA, Anderson TW, Newcombe HB, Lowry RB (May 1988). "Genetic disorders in children and young adults: a population study". American Journal of Human Genetics. 42 (5): 677–93. PMC 1715177. PMID 3358420.
- Scheffer PG, van der Schoot CE, Page-Christiaens GC, Bossers B, van Erp F, de Haas M (January 2010). "Reliability of fetal sex determination using maternal plasma". Obstetrics and Gynecology. 115 (1). Ovid Technologies (Wolters Kluwer Health): 117–26. doi:10.1097/aog.0b013e3181c3c938. PMID 20027043. S2CID 26126381.
- ^ Bustamante-Aragones A, Gonzalez-Gonzalez C, de Alba MR, Ainse E, Ramos C (March 2010). "Noninvasive prenatal diagnosis using ccffDNA in maternal blood: state of the art". Expert Review of Molecular Diagnostics. 10 (2). Informa UK Limited: 197–205. doi:10.1586/erm.09.86. PMID 20214538. S2CID 207219250.
- Zimmermann B, El-Sheikhah A, Nicolaides K, Holzgreve W, Hahn S (September 2005). "Optimized real-time quantitative PCR measurement of male fetal DNA in maternal plasma". Clinical Chemistry. 51 (9). American Association for Clinical Chemistry (AACC): 1598–604. doi:10.1373/clinchem.2005.051235. PMID 16020496.
- Finning KM, Chitty LS (April 2008). "Non-invasive fetal sex determination: impact on clinical practice". Seminars in Fetal & Neonatal Medicine. 13 (2). Elsevier BV: 69–75. doi:10.1016/j.siny.2007.12.007. PMID 18243829.
- Markey CM, Wadia PR, Rubin BS, Sonnenschein C, Soto AM (June 2005). "Long-term effects of fetal exposure to low doses of the xenoestrogen bisphenol-A in the female mouse genital tract". Biology of Reproduction. 72 (6). Oxford University Press (OUP): 1344–51. doi:10.1095/biolreprod.104.036301. PMID 15689538.
- Sayres LC, Cho MK (July 2011). "Cell-free fetal nucleic acid testing: a review of the technology and its applications". Obstetrical & Gynecological Survey. 66 (7). Ovid Technologies (Wolters Kluwer Health): 431–42. doi:10.1097/ogx.0b013e31822dfbe2. PMID 21944155. S2CID 17018886.
- Hill M, Barrett AN, White H, Chitty LS (October 2012). "Uses of cell free fetal DNA in maternal circulation". Best Practice & Research. Clinical Obstetrics & Gynaecology. 26 (5). Elsevier BV: 639–54. doi:10.1016/j.bpobgyn.2012.03.004. PMID 22542961.
- Norbury G, Norbury CJ (April 2008). "Non-invasive prenatal diagnosis of single gene disorders: how close are we?". Seminars in Fetal & Neonatal Medicine. 13 (2). Elsevier BV: 76–83. doi:10.1016/j.siny.2007.12.008. PMID 18234572.
- ^ Li Y, Page-Christiaens GC, Gille JJ, Holzgreve W, Hahn S (January 2007). "Non-invasive prenatal detection of achondroplasia in size-fractionated cell-free DNA by MALDI-TOF MS assay". Prenatal Diagnosis. 27 (1). Wiley-Blackwell: 11–7. doi:10.1002/pd.1608. PMID 17154237. S2CID 5808436.
- de Die-Smulders CE, de Wert GM, Liebaers I, Tibben A, Evers-Kiebooms G (2013). "Reproductive options for prospective parents in families with Huntington's disease: clinical, psychological and ethical reflections". Human Reproduction Update. 19 (3): 304–15. doi:10.1093/humupd/dms058. PMID 23377865.
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR (October 2008). "Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood". Proceedings of the National Academy of Sciences of the United States of America. 105 (42). Proceedings of the National Academy of Sciences: 16266–71. Bibcode:2008PNAS..10516266F. doi:10.1073/pnas.0808319105. PMC 2562413. PMID 18838674.
- ^ Cardo L, García BP, Alvarez FV (August 2010). "Non-invasive fetal RHD genotyping in the first trimester of pregnancy". Clinical Chemistry and Laboratory Medicine. 48 (8). Walter de Gruyter GmbH: 1121–6. doi:10.1515/cclm.2010.234. PMID 20482298. S2CID 31027958.
- Chinen PA, Nardozza LM, Martinhago CD, Camano L, Daher S, Pares DB, Minett T, Araujo Júnior E, Moron AF (November 2010). "Noninvasive determination of fetal rh blood group, D antigen status by cell-free DNA analysis in maternal plasma: experience in a Brazilian population". American Journal of Perinatology. 27 (10). Georg Thieme Verlag KG: 759–62. doi:10.1055/s-0030-1253560. PMID 20408112. S2CID 25705372.
- Okwundu CI, Afolabi BB (January 2013). "Intramuscular versus intravenous anti-D for preventing Rhesus alloimmunization during pregnancy". The Cochrane Database of Systematic Reviews (1): CD007885. doi:10.1002/14651858.CD007885.pub2. PMID 23440818.
- Aykut A, Onay H, Sagol S, Gunduz C, Ozkinay F, Cogulu O (December 2013). "Determination of fetal rhesus d status by maternal plasma DNA analysis". Balkan Journal of Medical Genetics. 16 (2). Walter de Gruyter GmbH: 33–8. doi:10.2478/bjmg-2013-0029. PMC 4001413. PMID 24778561.
- Svobodová I, Pazourková E, Hořínek A, Novotná M, Calda P, Korabečná M (2015). "Performance of Droplet Digital PCR in Non-Invasive Fetal RHD Genotyping - Comparison with a Routine Real-Time PCR Based Approach". PLOS ONE. 10 (11): e0142572. Bibcode:2015PLoSO..1042572S. doi:10.1371/journal.pone.0142572. PMC 4642940. PMID 26562517.
- Papasavva T, Martin P, Legler TJ, Liasides M, Anastasiou G, Christofides A, Christodoulou T, Demetriou S, Kerimis P, Kontos C, Leontiades G, Papapetrou D, Patroclos T, Phylaktou M, Zottis N, Karitzie E, Pavlou E, Kountouris P, Veldhuisen B, van der Schoot E, Kleanthous M (April 2016). "Prevalence of RhD status and clinical application of non-invasive prenatal determination of fetal RHD in maternal plasma: a 5 year experience in Cyprus". BMC Research Notes. 9 (1). Springer Nature: 198. doi:10.1186/s13104-016-2002-x. PMC 4818414. PMID 27036548.
- Zhang B, Lu BY, Yu B, Zheng FX, Zhou Q, Chen YP, Zhang XQ (April 2017). "Noninvasive prenatal screening for fetal common sex chromosome aneuploidies from maternal blood". The Journal of International Medical Research. 45 (2). SAGE Publications: 621–630. doi:10.1177/0300060517695008. PMC 5536640. PMID 28357876.
- Diagram by Mikael Häggström, MD, using following source: Jacquelyn V Halliday, MSGeralyn M Messerlian, PhDGlenn E Palomaki, PhD. "Patient education: Should I have a screening test for Down syndrome during pregnancy? (Beyond the Basics)". UpToDate.
{{cite web}}: CS1 maint: multiple names: authors list (link) This topic last updated: Feb 16, 2023. - ^ Kazemi M, Salehi M, Kheirollahi M (10 August 2016). "Down Syndrome: Current Status, Challenges and Future Perspectives". International Journal of Molecular and Cellular Medicine. 5 (3): 125–133. PMC 5125364. PMID 27942498.
- Mersy E, Smits LJ, van Winden LA, de Die-Smulders CE, Paulussen AD, Macville MV, Coumans AB, Frints SG (2013). "Noninvasive detection of fetal trisomy 21: systematic review and report of quality and outcomes of diagnostic accuracy studies performed between 1997 and 2012". Human Reproduction Update. 19 (4): 318–29. doi:10.1093/humupd/dmt001. PMID 23396607.
- Clark-Ganheart CA, Iqbal SN, Brown DL, Black S, Fries MH (May 2014). "Understanding the Limitations of Circulating Cell Free Fetal DNA: An Example of Two Unique Cases". Journal of Clinical Gynecology and Obstetrics. 3 (2): 38–70. doi:10.14740/jcgo229w. PMC 4185925. PMID 25298847.
- Wataganara T, LeShane ES, Farina A, Messerlian GM, Lee T, Canick JA, Bianchi DW (February 2003). "Maternal serum cell-free fetal DNA levels are increased in cases of trisomy 13 but not trisomy 18". Human Genetics. 112 (2): 204–8. doi:10.1007/s00439-002-0853-9. PMID 12522563. S2CID 9721963.
- Chiu RW, Lo YM (April 2011). "Non-invasive prenatal diagnosis by fetal nucleic acid analysis in maternal plasma: the coming of age". Seminars in Fetal & Neonatal Medicine. 16 (2). Elsevier BV: 88–93. doi:10.1016/j.siny.2010.10.003. PMID 21075065.
- Noninvasive Prenatal Diagnosis of Fetal Aneuploidy Using Cell-Free Fetal Nucleic Acids in Maternal Blood: Clinical Policy (Effective 05/01/2013) Archived 2014-03-07 at the Wayback Machine from Oxford Health Plans
- Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, Craig JA, Chudova DI, Devers PL, Jones KW, Oliver K, Rava RP, Sehnert AJ (February 2014). "DNA sequencing versus standard prenatal aneuploidy screening". The New England Journal of Medicine. 370 (9): 799–808. doi:10.1056/NEJMoa1311037. PMID 24571752. S2CID 13278444.. A recent study in the New England Journal of Medicine demonstrated the feasibility of using NIPT in a low risk population.
- Henderson JT, Thompson JH, Burda BU, Cantor A (April 2017). "Preeclampsia Screening: Evidence Report and Systematic Review for the US Preventive Services Task Force". JAMA. 317 (16). American Medical Association (AMA): 1668–1683. doi:10.1001/jama.2016.18315. PMID 28444285. S2CID 205077025.
- ^ Seval MM, Karabulut HG, Tükün A, Koç A (2015). "Cell free fetal DNA in the plasma of pregnant women with preeclampsia". Clinical and Experimental Obstetrics & Gynecology. 42 (6): 787–91. doi:10.12891/ceog1982.2015. PMID 26753487. S2CID 20971322.
- Lo YM, Lau TK, Zhang J, Leung TN, Chang AM, Hjelm NM, Elmes RS, Bianchi DW (October 1999). "Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21". Clinical Chemistry. 45 (10): 1747–51. doi:10.1093/clinchem/45.10.1747. PMID 10508120.
- Leung TN, Zhang J, Lau TK, Chan LY, Lo YM (January 2001). "Increased maternal plasma fetal DNA concentrations in women who eventually develop preeclampsia". Clinical Chemistry. 47 (1): 137–9. doi:10.1093/clinchem/47.1.137. PMID 11148193.
- Zhong XY, Holzgreve W, Hahn S (2002). "The levels of circulatory cell free fetal DNA in maternal plasma are elevated prior to the onset of preeclampsia". Hypertension in Pregnancy. 21 (1). Informa UK Limited: 77–83. doi:10.1081/prg-120002911. PMID 12044339. S2CID 72519129.
- Ramezanzadeh, Mahboubeh; Khosravi, Sharifeh; Salehi, Rasoul (2017). "Cell-free Fetal Nucleic Acid Identifier Markers in Maternal Circulation". Advanced Biomedical Research. 6 (1): 89. doi:10.4103/2277-9175.211800. ISSN 2277-9175. PMC 5549546. PMID 28828340.
- Lo, Y M Dennis; Corbetta, Noemi; Chamberlain, Paul F; Rai, Vik; Sargent, Ian L; Redman, Christopher WG; Wainscoat, James S (1997-08-16). "Presence of fetal DNA in maternal plasma and serum". The Lancet. 350 (9076): 485–487. doi:10.1016/S0140-6736(97)02174-0. PMID 9274585.
- Hofschneider, Mark. "Noninvasive prenatal testing using fetal DNA". Lasker Foundation. Retrieved 2024-05-06.
- Yurkiewicz IR, Korf BR, Lehmann LS (January 2014). "Prenatal whole-genome sequencing--is the quest to know a fetus's future ethical?". The New England Journal of Medicine. 370 (3): 195–7. doi:10.1056/NEJMp1215536. PMID 24428465. S2CID 205109276.
- Wellcome Trust Case Control Consortium (June 2007). "Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls". Nature. 447 (7145): 661–78. Bibcode:2007Natur.447..661B. doi:10.1038/nature05911. PMC 2719288. PMID 17554300.
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, et al. (October 2007). "The NCBI dbGaP database of genotypes and phenotypes". Nature Genetics. 39 (10): 1181–6. doi:10.1038/ng1007-1181. PMC 2031016. PMID 17898773.