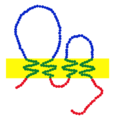
Claudins are a family of proteins which, along with occludin, are the most important components of the tight junctions (zonulae occludentes). Tight junctions establish the paracellular barrier that controls the flow of molecules in the intercellular space between the cells of an epithelium. They have four transmembrane domains, with the N-terminus and the C-terminus in the cytoplasm.
Structure
Claudins are small (20–24/27 kilodalton (kDa)) transmembrane proteins which are found in many organisms, ranging from nematodes to human beings. They all have a very similar structure. Claudins span the cellular membrane 4 times, with the N-terminal end and the C-terminal end both located in the cytoplasm, and two extracellular loops which show the highest degree of conservation.
Claudins have both cis and trans interactions between cell membranes. Cis-interactions is when claudins on the same membrane interact, one way they interact is by transmembrane domain having molecular interactions. Trans-interaction is when claudins of neighboring cells interact through their extracellular loops. Cis-interactions is also known as side-to-side interactions and trans-interactions is also known as head-to-head interactions.
Generally the tight junction is known for its impermeability. However, depending on the type of claudin and their interactions there is selective permeability. This includes charge selectivity and size selectivity.
N-terminal
The N-terminal end is usually very short (1–10 amino acids) It is located in the cytoplasm where it is thought to help to contribute to cell signaling, cytoskeletal organization and other possible functions.
C-terminal
The C-terminal has a longer chain and is located in the cytoplasm. It varies in length from 21 to 63 and is necessary for the localization of these proteins in the tight junctions. It is thought that it may play a role in cell signaling. All human claudins (with the exception of Claudin 12) have domains that let them bind to PDZ domains of scaffold proteins.
Transmembrane domain
The transmembrane domain is the amino acids that cross the cellular membrane. The transmembrane domain is important for cis-interaction of claudins.
First extracellular loop
The first extracellular loop has a range of 42-56 amino acids and is longer than the second extracellular loop. It is suspected that the cysteines of found on the first extracellular loop form disulfide bonds. This loop has charged amino acids that may be the predictor for the charge selectivity of tight junctions. The first extracellular loop plays a role in trans-interaction of claudins of adjacent cells.
Second extracellular loop
The second extracellular loop is shorter than the first extracellular loop. In this short chain of amino acids there are three hydrophobic residues. These three residues are suspected to be a contributor to the trans-interaction of proteins between adjacent cells.
History
Claudins were first named in 1998 by Japanese researchers Mikio Furuse and Shoichiro Tsukita at Kyoto University. The name claudin comes from Latin word claudere ("to close"), suggesting the barrier role of these proteins.
Studies
A recent review discusses evidence regarding the structure and function of claudin family proteins using a systems approach to understand evidence generated by proteomics techniques.
A chimeric claudin was synthesized to help enhance the understanding of both the structure and function of the tight junction.
Computational modeling is also another technique being used to help enhance research into the structure and functions of claudins.
Genes
| PMP22_Claudin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PMP22_Claudin | ||||||||
| Pfam | PF00822 | ||||||||
| Pfam clan | CL0375 | ||||||||
| InterPro | IPR004031 | ||||||||
| PROSITE | PDOC01045 | ||||||||
| TCDB | 1.H.1 | ||||||||
| OPM superfamily | 194 | ||||||||
| OPM protein | 4p79 | ||||||||
| |||||||||
There are 23 genes found in the human genome for claudin proteins and there are 27 transmembrane domains across mammals. The conservation is not observed on a genetic level. Despite the genetic level not being conserved across claudins their structural conservation are very similar.
- CLDN1, CLDN2, CLDN3, CLDN4, CLDN5, CLDN6, CLDN7, CLDN8, CLDN9, CLDN10, CLDN11, CLDN12, CLDN13, CLDN14, CLDN15, CLDN16, CLDN17, CLDN18, CLDN19, CLDN20, CLDN21, CLDN22, CLDN23
See also
Additional images
References
- ^ Hou J, Konrad M (2010-01-01). "Chapter 7 - Claudins and Renal Magnesium Handling". In Yu AS (ed.). Current Topics in Membranes. Vol. 65. Academic Press. pp. 151–176. doi:10.1016/s1063-5823(10)65007-7. ISBN 9780123810397.
- Furuse M (2010-01-01). "Chapter 1 - Introduction: Claudins, Tight Junctions, and the Paracellular Barrier". In Yu AS (ed.). Current Topics in Membranes. Vol. 65. Academic Press. pp. 1–19. doi:10.1016/s1063-5823(10)65001-6. ISBN 9780123810397.
- Szaszi K, Amoozadeh Y (2014-01-01). "Chapter Six - New Insights into Functions, Regulation, and Pathological Roles of Tight Junctions in Kidney Tubular Epithelium". In Jeon KW (ed.). International Review of Cell and Molecular Biology. Vol. 308. Academic Press. pp. 205–271. doi:10.1016/b978-0-12-800097-7.00006-3. ISBN 9780128000977. PMID 24411173.
- Otani T, Nguyen TP, Tokuda S, Sugihara K, Sugawara T, Furuse K, et al. (October 2019). "Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity". The Journal of Cell Biology. 218 (10): 3372–3396. doi:10.1083/jcb.201812157. PMC 6781433. PMID 31467165.
- Greene C, Campbell M, Janigro D (2019-01-01). "Chapter 1 - Fundamentals of Brain–Barrier Anatomy and Global Functions". In Lonser RR, Sarntinoranont M, Bankiewicz K (eds.). Nervous System Drug Delivery. Academic Press. pp. 3–20. doi:10.1016/b978-0-12-813997-4.00001-3. ISBN 978-0-12-813997-4. S2CID 198273920.
- Haseloff RF, Piontek J, Blasig IE (2010-01-01). "Chapter 5 - The Investigation of cis- and trans-Interactions Between Claudins". In Yu AS (ed.). Current Topics in Membranes. Vol. 65. Academic Press. pp. 97–112. doi:10.1016/s1063-5823(10)65005-3. ISBN 9780123810397.
- ^ Günzel D, Yu AS (April 2013). "Claudins and the modulation of tight junction permeability". Physiological Reviews. 93 (2): 525–569. doi:10.1152/physrev.00019.2012. PMC 3768107. PMID 23589827.
- ^ "Crystal Structures of claudins: insights into their intermolecular interactions". Annals of the New York Academy of Sciences. 1205. September 2010. doi:10.1111/nyas.2010.1205.issue-s1. ISSN 0077-8923.
- ^ Fuladi S, Jannat RW, Shen L, Weber CR, Khalili-Araghi F (January 2020). "Computational Modeling of Claudin Structure and Function". International Journal of Molecular Sciences. 21 (3): 742. doi:10.3390/ijms21030742. PMC 7037046. PMID 31979311.
- ^ Rüffer C, Gerke V (May 2004). "The C-terminal cytoplasmic tail of claudins 1 and 5 but not its PDZ-binding motif is required for apical localization at epithelial and endothelial tight junctions". European Journal of Cell Biology. 83 (4): 135–144. doi:10.1078/0171-9335-00366. PMID 15260435.
- ^ Tsukita S, Tanaka H, Tamura A (February 2019). "The Claudins: From Tight Junctions to Biological Systems". Trends in Biochemical Sciences. 44 (2): 141–152. doi:10.1016/j.tibs.2018.09.008. PMID 30665499. S2CID 58640701.
- Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S (June 1998). "Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin". The Journal of Cell Biology. 141 (7): 1539–1550. doi:10.1083/jcb.141.7.1539. PMC 2132999. PMID 9647647.
- Liu F, Koval M, Ranganathan S, Fanayan S, Hancock WS, Lundberg EK, et al. (February 2016). "Systems Proteomics View of the Endogenous Human Claudin Protein Family". Journal of Proteome Research. 15 (2): 339–359. doi:10.1021/acs.jproteome.5b00769. PMC 4777318. PMID 26680015.
- ^ Taylor A, Warner M, Mendoza C, Memmott C, LeCheminant T, Bailey S, et al. (May 2021). "Chimeric Claudins: A New Tool to Study Tight Junction Structure and Function". International Journal of Molecular Sciences. 22 (9): 4947. doi:10.3390/ijms22094947. PMC 8124314. PMID 34066630.
- Hou J (2019-01-01). "Chapter 7 - Paracellular Channel in Organ System". In Hou J (ed.). The Paracellular Channel. Academic Press. pp. 93–141. doi:10.1016/b978-0-12-814635-4.00007-3. ISBN 978-0-12-814635-4. S2CID 90477792.
- Hou J (2019-01-01). "Chapter 8 - Paracellular Channel in Human Disease". In Hou J (ed.). The Paracellular Channel. Academic Press. pp. 143–173. doi:10.1016/b978-0-12-814635-4.00008-5. ISBN 978-0-12-814635-4. S2CID 90122806.
| Proteins of epithelium | |
|---|---|
| Lateral/cell–cell | |
| Basal/cell–matrix | |
| Apical | |