| Crotamine | |||||||
|---|---|---|---|---|---|---|---|
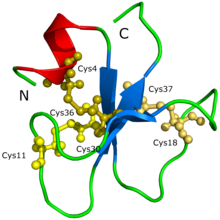 The protein structure of crotamine (PDB: 1H5O) The protein structure of crotamine (PDB: 1H5O) | |||||||
| Identifiers | |||||||
| Organism | ? | ||||||
| Symbol | CRO2 | ||||||
| UniProt | Q9PWF3 | ||||||
| |||||||
Crotamine is a toxin present in the venom of the South American rattlesnake (Crotalus durissus terrificus). It is a 42-residue-long protein containing 11 basic residues (nine lysines, two arginines) and six cysteines. It has also been isolated from the venom of North American prairie rattlesnake, Crotalus viridis viridis. It was first isolated and purified by Brazilian scientist José Moura Gonçalves, and later intensively studied by his group of collaborators at the Medical School of Ribeirão Preto of the University of São Paulo.
Biological function
Crotamine has a number of biological actions: it acts on cell membrane's sodium channels, is slightly analgesic and is myotoxic, i.e., it penetrates the cells of muscles and promotes necrosis. Crotamine is homologous with other venom myotoxins and is similar to α-,β-defensins.
Biochemistry and mechanism
The amino acid sequence (YKQCHKKGGHCFPKEKICLPPSSDFGKMDCRWRWKCCKKGSG, Cys4-Cys36, Cys11-Cys30, Cys18-Cys37) and the 3D molecular structure of crotamine have already been determined.
The protein structure of crotamine could not be initially determined through protein crystallization nor X-ray diffraction. It was speculated that the difficulty was because crotamine has so many isoforms, leading to the formation of aggregates and different possible conformations of the protein. The structure and the shape of the protein was proposed through a 3D model generated by Siqueira et al. (2002) based on computational calculations that were supported with intensive molecular dynamics simulations and homology modeling procedures. Afterwards, Nicastro et al. (2003) discovered the structure of crotamine through nuclear magnetic resonance spectroscopy. Crotamine has a topology that was never before seen in active toxins that target ion channels; the protein is composed of a short N-terminal alpha helix, a type of protein formation, and a small antiparallel triple-stranded beta-sheet, another type of protein formation, arranged in an ab1b2b3 topology. Crotamine has similar structural fold conformations to the human β-defensin family as well as an identical arrangement of three disulfide bridges.
The gene and chromosome location responsible for its synthesis have been identified by the group led by Gandhi Rádis-Baptista, working at the Instituto Butantan, in São Paulo, Brazil. The mRNA has about 340 nucleotides and codifies a pre-crotamine, including the signal peptide, the mature crotamine, and a final lysine.
The Crotamine gene was the first gene to be mapped on a snake chromosome. The gene responsible for coding the crotamine protein is labeled as Crt-p1 and its base pair sequence length is about 1.1kbp or 1100 bp. It was reported that the crotamine gene was isolated twice from two different specimens, one in a method that resulted in a gene size of 1.8 kbp and in the other specimen a gene size of 1.1 kbp. The gene has been previously isolated in the C. durissus terrificus genome and the protein itself belongs to a group of small basic polypeptide myotoxins (SBPM). The contents of Crotalus venoms can vary according to subspecies and geographical location. The Crt-p1 gene, as described by Radis-Bastista et al. 2003, consists of about three exons separated by one short phase-2 (140 bp) and one long phase-1 (900 bp) intron. Exon 1 codes for the first 19 amino acids of the signal peptide and includes the 5’-untranslated region. Exon 2 codes 39 amino acids to the mature crotamine and three signal peptide amino acids. Exon 3 codes for the terminal lysine and the last three amino acids of the mature toxin. Research on SBPM amino acid sequences among different Crotalus species has revealed a high degree of likeness ranging from 83% - 98%.
The amino acid code of proteins in the small basic polypeptide myotoxin family, which includes crotamine, have been sequenced. They were found to be similar with an average of 83% divergence. A crotamine amino acid sequence was compared to that of cloned DNA of myotoxin a, (the myotoxin used to model how SBPMs work). In the comparison, exon coding regions including the mature myotoxin and the signal peptide were 98% and 100% similar, respectively. The untranslated regions for 5’ and 3’ between the sample and the myotoxin a cDNA was 60% and 80%, respectively. When comparing the amino acid sequences of other proteins not in the SBPM family found in snake venoms, there is usually large divergence. When looking at the SBPM proteins, they have high similarity between different subspecies of the Crotalus genus and between different individuals of the same subspecies. This indicates, according to the Radis-Batista et al. 2003 study, that the crotamine gene and other SBPM genes have evolved recently.
References
- Oliveira AL, Viegas MF, da Silva SL, Soares AM, Ramos MJ, Fernandes PA (2022-06-10). "The chemistry of snake venom and its medicinal potential". Nature Reviews. Chemistry. 6 (7): 451–469. doi:10.1038/s41570-022-00393-7. PMC 9185726. PMID 35702592.
- ^ Oguiura N, Boni-Mitake M, Rádis-Baptista G (September 2005). "New view on crotamine, a small basic polypeptide myotoxin from South American rattlesnake venom". Toxicon. 46 (4): 363–370. doi:10.1016/j.toxicon.2005.06.009. PMID 16115660. S2CID 40142037.
- ^ Samejima Y, Aoki Y, Mebs D (1991). "Amino acid sequence of a myotoxin from venom of the eastern diamondback rattlesnake (Crotalus adamanteus)". Toxicon. 29 (4–5): 461–468. doi:10.1016/0041-0101(91)90020-r. PMID 1862521.
- Schenberg S (May 1959). "Geographical pattern of crotamine distribution in the same rattlesnake subspecies". Science. 129 (3359): 1361–1363. doi:10.1126/science.129.3359.1361. PMID 13658964.
Further reading
- Goncalves JM, Deutsch HF (February 1956). "Ultracentrifugal and zone electrophoresis studies of some crotalidae venoms". Archives of Biochemistry and Biophysics. 60 (2): 402–411. doi:10.1016/0003-9861(56)90444-1. PMID 13292919.
- Giglio JR (November 1975). "Analytical studies on crotamine hydrochloride". Analytical Biochemistry. 69 (1): 207–221. doi:10.1016/0003-2697(75)90581-3. PMID 2030.
- Laure CJ (February 1975). "". Hoppe-Seyler's Zeitschrift für Physiologische Chemie (in German). 356 (2): 213–215. PMID 1176086.
- De Lucca FL, Imaizumi MT, Haddad A (April 1974). "Characterization of ribonucleic acids from the venom glands of Crotalus durissus terrifucus (Ophidia, Reptilia) after manual extraction of the venom. Studies on template activity and base composition". The Biochemical Journal. 139 (1): 151–156. doi:10.1042/bj1390151. PMC 1166261. PMID 4463939.
- Ownby CL, Cameron D, Tu AT (October 1976). "Isolation of myotoxic component from rattlesnake (Crotalus viridis viridis) venom. Electron microscopic analysis of muscle damage". The American Journal of Pathology. 85 (1): 149–166. PMC 2032543. PMID 970437.
- Rádis-Baptista G, Oguiura N, Hayashi MA, Camargo ME, Grego KF, Oliveira EB, Yamane T (July 1999). "Nucleotide sequence of crotamine isoform precursors from a single South American rattlesnake (Crotalus durissus terrificus)". Toxicon. 37 (7): 973–984. doi:10.1016/s0041-0101(98)00226-8. PMID 10484745.
- Kerkis A, Kerkis I, Rádis-Baptista G, Oliveira EB, Vianna-Morgante AM, Pereira LV, Yamane T (September 2004). "Crotamine is a novel cell-penetrating protein from the venom of rattlesnake Crotalus durissus terrificus". FASEB Journal. 18 (12): 1407–1409. doi:10.1096/fj.03-1459fje. PMID 15231729. S2CID 20510076.
- Rádis-Baptista G, Kubo T, Oguiura N, Prieto da Silva AR, Hayashi MA, Oliveira EB, Yamane T (June 2004). "Identification of crotasin, a crotamine-related gene of Crotalus durissus terrificus". Toxicon. 43 (7): 751–759. doi:10.1016/j.toxicon.2004.02.023. PMID 15284009. S2CID 16350991.
- Rádis-Baptista G, Kubo T, Oguiura N, Svartman M, Almeida TM, Batistic RF, et al. (December 2003). "Structure and chromosomal localization of the gene for crotamine, a toxin from the South American rattlesnake, Crotalus durissus terrificus". Toxicon. 42 (7): 747–752. doi:10.1016/j.toxicon.2003.10.019. PMID 14757205.
- Nicastro G, Franzoni L, de Chiara C, Mancin AC, Giglio JR, Spisni A (May 2003). "Solution structure of crotamine, a Na+ channel affecting toxin from Crotalus durissus terrificus venom". European Journal of Biochemistry. 270 (9): 1969–1979. doi:10.1046/j.1432-1033.2003.03563.x. PMID 12709056. S2CID 20601072.
- Mouhat S, Jouirou B, Mosbah A, De Waard M, Sabatier JM (March 2004). "Diversity of folds in animal toxins acting on ion channels". The Biochemical Journal. 378 (Pt 3): 717–726. doi:10.1042/BJ20031860. PMC 1224033. PMID 14674883.
External links
- Nucleotide sequence and translation for crotasin. Entrez Database. National Center for Biotechnology Information.