| |
| Names | |
|---|---|
| IUPAC name 7H-purine-2,6-diamine | |
| Other names 2-aminoadenine; 2,6-DAP | |
| Identifiers | |
| CAS Number | |
| 3D model (JSmol) | |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.016.006 |
| PubChem CID | |
| UNII | |
| CompTox Dashboard (EPA) | |
InChI
| |
SMILES
| |
| Properties | |
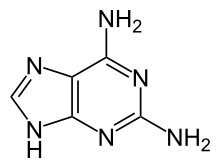
| Chemical formula | C5H6N6 |
| Molar mass | 150.145 g·mol |
| Appearance | White to yellow crystalline powder |
| Density | 1.743 g/cm |
| Melting point | 117 to 122 °C (243 to 252 °F; 390 to 395 K) |
| Solubility in water | 2.38 g/L at 20 °C |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C , 100 kPa).
| |
2,6-diaminopurine (2,6-DAP, also known as 2-aminoadenine) is a compound once used in the treatment of leukemia. As the Z base, it is found instead of adenine (A) in the genetic material of some bacteriophage viruses.
In August 2011, a report, based on NASA studies with meteorites found on Earth, was published suggesting 2,6-diaminopurine and related organic molecules, including the DNA and RNA components adenine and guanine, may have been formed extraterrestrially in outer space.
In viruses
Main article: nucleic acid analogues
In cyanophage S-2L (Siphoviridae), diaminopurine is used instead of adenine (host evasion). Diaminopurine base (Z) pairs perfectly with thymine (T) as it is identical to adenine (A) but has an amine group at position 2 forming 3 intermolecular hydrogen bonds, eliminating the major difference between the two types of basepairs (weak:A-T and strong:C-G). This improved stability affects protein-binding interactions that rely on those differences.
Four papers published April 2021 further describes the use and production of the Z-base. It is now known that:
- The S-2L phage avoids incorporating A bases in the genome by hydrolyzing dATP (DatZ enzyme);
- The Z base is produced by a pathway involving DUF550 (MazZ) and PurZ in S-2L and Vibrio phage PhiVC8;
- The PrimPol/AEP DNA polymerase responsible for handling the Z base occurs in the same gene cluster as the three aforementioned enzymes;
- The Z base is quite widespread in both Siphoviridae and Podoviridae, based on the occurrence of the said gene cluster.
In August 2021, it was shown that DatZ, MazZ and PurZ are sufficient to replace some occurrence of A by Z in the bacterial genome of E. coli; expression of this system is toxic to the cell. The structures of MazZ (subtype 2) and PurZ are also determined, showing a possible link between PurZ and archaeal versions of PurA.
Biosynthesis
2-aminoadenine is produced in two steps. The enzyme MazZ (homologous to MazG, EC 3.6.1.8) first performs:
- dGTP + H2O = dGMP + diphosphate
The enzyme PurZ (homologous to PurA, EC 6.3.4.4) then performs:
- (d)ATP + dGMP + L-aspartate = (d)ADP + phosphate + 2-aminodeoxyadenylosuccinate (dSMP)
The resulting dSMP is processed by host enzymes analogously to adenylosuccinate to produce dZTP.
In cellular life
| This article is missing information about results of the altered H-bond strength in DNA and RNA. Please expand the article to include this information. Further details may exist on the talk page. (October 2021) |
2,6-DAP was used to treat leukemia since as early as 1951. It is known to arrest progression of cell cycle in mouse leukemia cells by 1989. Cancer cells are known to become resistant to DAP by losing their adenine phosphoribosyltransferase (APRT) function, a process shared with E. coli.
DAP derivatives are in vitro antivirals useful against pseudorabies virus, a economically important livestock disease. This base, in its free form, is able to correct UGA nonsense mutations by encouraging translational readthrough, through the inhibition of FTSJ1.
Bioengineering
In bioengineering, anti-miRNA oligonucleotides (specifically, the serinol nucleic acid type) incorporating base Z instead of A show enhanced binding to RNA.
DAP is used similarly to other nuclear acid analogues in the investigation of enzyme structures and mechanisms.
References
- "George H. Hitchings". nobelprize.org.
- "Some viruses thwart bacterial defenses with a unique genetic alphabet". 5 May 2021.
- Callahan, M.P.; Smith, K.E.; Cleaves, H.J.; Ruzica, J.; Stern, J.C.; Glavin, D.P.; House, C.H.; Dworkin, J.P. (11 August 2011). "Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases". Proceedings of the National Academy of Sciences. 108 (34). PNAS: 13995–13998. Bibcode:2011PNAS..10813995C. doi:10.1073/pnas.1106493108. PMC 3161613. PMID 21836052.
- Steigerwald, John (8 August 2011). "NASA Researchers: DNA Building Blocks Can Be Made in Space". NASA. Retrieved 2011-08-10.
- ScienceDaily Staff (9 August 2011). "DNA Building Blocks Can Be Made in Space, NASA Evidence Suggests". ScienceDaily. Retrieved 2011-08-09.
- Kirnos, MD; Khudyakov, IY; Alexandrushkina, NI; Vanyushin, BF (November 1977). "2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA". Nature. 270 (5635): 369–70. Bibcode:1977Natur.270..369K. doi:10.1038/270369a0. PMID 413053.
- Bowler, Jacinta (4 May 2021). "Some Viruses Have a Completely Different Genome to The Rest of Life on Earth". ScienceAlert.
- Czernecki, Dariusz; Legrand, Pierre; Tekpinar, Mustafa; Rosario, Sandrine; Kaminski, Pierre-Alexandre; Delarue, Marc (2021-04-23). "How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA". Nature Communications. 12 (1): 2420. Bibcode:2021NatCo..12.2420C. doi:10.1038/s41467-021-22626-x. ISSN 2041-1723. PMC 8065100. PMID 33893297.
- ^ Sleiman, Dona; Garcia, Pierre Simon; Lagune, Marion; Loc’h, Jerome; Haouz, Ahmed; Taib, Najwa; Röthlisberger, Pascal; Gribaldo, Simonetta; Marlière, Philippe; Kaminski, Pierre Alexandre (30 April 2021). "A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes". Science. 372 (6541): 516–520. Bibcode:2021Sci...372..516S. doi:10.1126/science.abe6494. PMID 33926955. S2CID 233448787.
- Pezo, Valerie; Jaziri, Faten; Bourguignon, Pierre-Yves; Louis, Dominique; Jacobs-Sera, Deborah; Rozenski, Jef; Pochet, Sylvie; Herdewijn, Piet; Hatfull, Graham F.; Kaminski, Pierre-Alexandre; Marliere, Philippe (30 April 2021). "Noncanonical DNA polymerization by aminoadenine-based siphoviruses". Science. 372 (6541): 520–524. Bibcode:2021Sci...372..520P. doi:10.1126/science.abe6542. PMID 33926956. S2CID 233448788.
- Zhou, Yan; Su, Xuexia; Wei, Yifeng; Cheng, Yu; Guo, Yu; Khudyakov, Ivan; Liu, Fuli; He, Ping; Song, Zhangyue; Li, Zhi; Gao, Yan; Ang, Ee Lui; Zhao, Huimin; Zhang, Yan; Zhao, Suwen (30 April 2021). "A widespread pathway for substitution of adenine by diaminopurine in phage genomes". Science. 372 (6541): 512–516. Bibcode:2021Sci...372..512Z. doi:10.1126/science.abe4882. PMID 33926954. S2CID 233448821.
- ^ Czernecki, Dariusz; Bonhomme, Frédéric; Kaminski, Pierre-Alexandre; Delarue, Marc (2021-08-05). "Characterization of a triad of genes in cyanophage S-2L sufficient to replace adenine by 2-aminoadenine in bacterial DNA". Nature Communications. 12 (1): 4710. Bibcode:2021NatCo..12.4710C. bioRxiv 10.1101/2021.04.30.442174. doi:10.1038/s41467-021-25064-x. ISSN 2041-1723. PMC 8342488. PMID 34354070.
- Burchenal, JH; Karnofsky, DA; Kingsley-Pillers, EM; Southam, CM; Myers, WP; Escher, GC; Craver, LF; Dargeon, HW; Rhoads, CP (May 1951). "The effects of the folic acid antagonists and 2,6-diaminopurine on neoplastic disease, with special reference to acute leukemia". Cancer. 4 (3): 549–69. doi:10.1002/1097-0142(195105)4:3<549::aid-cncr2820040308>3.0.co;2-j. PMID 14839611. S2CID 31262125.
- Weckbecker, G; Cory, JG (1989). "Metabolic activation of 2,6-diaminopurine and 2,6-diaminopurine-2'-deoxyriboside to antitumor agents". Advances in Enzyme Regulation. 28: 125–44. doi:10.1016/0065-2571(89)90068-x. PMID 2624171.
- Shao, C; Deng, L; Henegariu, O; Liang, L; Stambrook, PJ; Tischfield, JA (20 June 2000). "Chromosome instability contributes to loss of heterozygosity in mice lacking p53". Proceedings of the National Academy of Sciences of the United States of America. 97 (13): 7405–10. Bibcode:2000PNAS...97.7405S. doi:10.1073/pnas.97.13.7405. PMC 16558. PMID 10861008.
- Kocharian, ShM; Chukanova, TI; Sukhodolets, VV (1977). "". Genetika. 13 (10): 1821–30. PMID 348574.
- Zouharova, D; Lipenska, I; Fojtikova, M; Kulich, P; Neca, J; Slany, M; Kovarcik, K; Turanek-Knotigova, P; Hubatka, F; Celechovska, H; Masek, J; Koudelka, S; Prochazka, L; Eyer, L; Plockova, J; Bartheldyova, E; Miller, AD; Ruzek, D; Raska, M; Janeba, Z; Turanek, J (29 February 2016). "Antiviral activities of 2,6-diaminopurine-based acyclic nucleoside phosphonates against herpesviruses: In vitro study results with pseudorabies virus (PrV, SuHV-1)". Veterinary Microbiology. 184: 84–93. doi:10.1016/j.vetmic.2016.01.010. PMID 26854349.
- Trzaska, C; Amand, S; Bailly, C; Leroy, C; Marchand, V; Duvernois-Berthet, E; Saliou, JM; Benhabiles, H; Werkmeister, E; Chassat, T; Guilbert, R; Hannebique, D; Mouray, A; Copin, MC; Moreau, PA; Adriaenssens, E; Kulozik, A; Westhof, E; Tulasne, D; Motorin, Y; Rebuffat, S; Lejeune, F (20 March 2020). "2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations". Nature Communications. 11 (1): 1509. Bibcode:2020NatCo..11.1509T. doi:10.1038/s41467-020-15140-z. PMC 7083880. PMID 32198346.
- Kamiya, Y; Donoshita, Y; Kamimoto, H; Murayama, K; Ariyoshi, J; Asanuma, H (5 October 2017). "Introduction of 2,6-Diaminopurines into Serinol Nucleic Acid Improves Anti-miRNA Performance". ChemBioChem. 18 (19): 1917–1922. doi:10.1002/cbic.201700272. PMID 28748559. S2CID 35619213.
- Bailly, C (1 October 1998). "The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA". Nucleic Acids Research. 26 (19): 4309–4314. doi:10.1093/nar/26.19.4309. PMC 147870. PMID 9742229.