| formyl peptide receptor 1 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | FPR1 | ||||||
| Alt. symbols | FPR; FMLP | ||||||
| NCBI gene | 2357 | ||||||
| HGNC | 3826 | ||||||
| OMIM | 136537 | ||||||
| RefSeq | NM_002029 | ||||||
| UniProt | P21462 | ||||||
| Other data | |||||||
| Locus | Chr. 19 q13.41 | ||||||
| |||||||
| formyl peptide receptor 2 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | FPR2 | ||||||
| Alt. symbols | ALXR, FMLPX, FPR2/ALX, FPR2A, FPRH1, FPRL1, HM63, LXA4R, RFP | ||||||
| NCBI gene | 2358 | ||||||
| HGNC | 3827 | ||||||
| OMIM | 136538 | ||||||
| RefSeq | NM_001462 | ||||||
| UniProt | P25090 | ||||||
| Other data | |||||||
| Locus | Chr. 19 q13.3-13.4 | ||||||
| |||||||
| formyl peptide receptor 3 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | FPR3 | ||||||
| Alt. symbols | FPRH2, FPRL2, FMLPY | ||||||
| NCBI gene | 2359 | ||||||
| HGNC | 3828 | ||||||
| OMIM | 136539 | ||||||
| RefSeq | NM_002030 | ||||||
| UniProt | P25089 | ||||||
| Other data | |||||||
| Locus | Chr. 19 q13.3-13.4 | ||||||
| |||||||
The formyl peptide receptors (FPR) belong to a class of G protein-coupled receptors involved in chemotaxis. In humans, there are three formyl peptide receptor isoforms, each encoded by a separate gene that are named FPR1, FPR2, and FPR3. These receptors were originally identified by their ability to bind N-formyl peptides such as N-formylmethionine produced by the degradation of either bacterial or host cells. Hence formyl peptide receptors are involved in mediating immune cell response to infection. These receptors may also act to suppress the immune system under certain conditions. The close phylogenetic relation of signaling in chemotaxis and olfaction was recently proved by detection formyl peptide receptor like proteins as a distinct family of vomeronasal organ chemosensors in mice.
FPR is now properly accepted as termed FPR1 by the International Union of Basic and Clinical Pharmacology.
Discovery
Studies conducted in the 1970s found that a series of N-Formylmethionine-containing oligopeptides, including the most potent and best known member of this series, N-formylmethionine-leucyl-phenylalanine (fMLF or fMet-Leu-Phe), stimulated rabbit and human neutrophils by an apparent receptor-dependent mechanism to migrate in a directional pattern in classical laboratory assays of chemotaxis. Since these oligopeptides were produced by bacteria or synthetic analogs of such products, it was suggested that the N-formyl oligopeptides are important chemotatic factors and their receptors are important chemotactic factor receptors that act respectively as signaling and signal-recognizing elements to initiate inflammation responses in order to defend against bacterial invasion. Further studies defined a receptor for the N-formyl oligopeptides, formyl peptide receptor (FPR), so named based on its ability to bind and become activated by the oligopeptides. Two receptors where thereafter discovered and named FPR1 and FPR2 based on the similarity of their genes' predicted amino acid sequence to that of FPR rather than on any ability to bind or be activated by the formyl oligopeptides. These three receptors have been renamed as FPR1, FPR2, and FPR3 and found to have very different specificities for the formyl oligopeptides and very different functions that include initiating inflammatory responses to N-formyl peptides released not only by bacteria but also a multiplicity of elements released by host tissues; dampening and resolving inflammatory responses; and perhaps contributing to the development of certain neurological cancers and an array of neurological diseases Amyloid-based diseases.
Structure and function
The formyl peptide receptor (FPR) belongs to the class of receptors possessing seven hydrophobic transmembrane domains. The conformation of the FPR is stabilized by several interactions. These include potential salt bridge formation between Arg84-Arg205, Lys85-Arg205, and Lys85-Asp284 which help determine the three-dimensional structure of transmembrane domains, as well as positively charged residues (Arg, Lys) which interact with negatively charged phosphates. Furthermore, residue Arg163 may interact with the ligand binding pocket of the second extracellular loop of the FPR.
With respect to binding of the formyl Met-Leu-Phe peptide, there are additional potential interactions which include hydrogen bonding interactions between Arg84 and Lys85 of the first extracellular loop and the N-formyl group of the ligand as well as the peptide backbone of formyl Met-Leu-Phe which can form similar interactions. The formyl-Met moiety of the ligand was shown to form disulfide bridges with Cys residues, and an interaction with Arg163 was also demonstrated. (It is important to mention that some interaction which stabilize the conformation of the receptor may also influence ligand-binding.) Some oligopeptides were also described as characteristic constituents linked to Asn-s of the extracellular N terminal part and to the ligand binding pocket of the second extracellular loop. These components can also determine or make more specific the ligand-receptor interaction.
-
 Schematic diagram of the formyl peptide receptor 1. Transmembrane helices of the receptor are represented by blue-green cylinders while the cell membrane in which the receptor is imbedded is depicted in yellow. The extracellular face of the cell membrane is on top while the intracellular (cytoplasmic) face is on the bottom. Extracellular loops of the FPR responsible for N-for-Met-Leu-Phe (Nfor-MLF) binding are shown in red.
Schematic diagram of the formyl peptide receptor 1. Transmembrane helices of the receptor are represented by blue-green cylinders while the cell membrane in which the receptor is imbedded is depicted in yellow. The extracellular face of the cell membrane is on top while the intracellular (cytoplasmic) face is on the bottom. Extracellular loops of the FPR responsible for N-for-Met-Leu-Phe (Nfor-MLF) binding are shown in red.
-
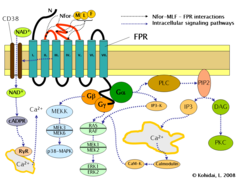 Formyl peptide receptor (FPR) signaling pathways.
Formyl peptide receptor (FPR) signaling pathways.
Signaling pathways
Induction of FPR triggers multiple changes in eukaryotic cells including rearrangement of the cytoskeleton which in turn facilitates cell migration and the synthesis of chemokines. Important FPR regulated pathways include:
- G protein dependent activation of phospholipase C (PLC) which results in the breakdown of the membrane constituent phospholipid, phosphatidylinositol (4,5)-bisphosphate (PIP2) into inositol (1,4,5)-trisphosphate (IP3) and diacyl glycerol (DAG). IP3 is one of the most effective inducers of Ca increase from cytoplasmic pools and from outside the cell via opening Ca channels. DAG in turn is an inducer of protein kinase C (PKC).
- Activation of the regulatory small GTPase, RAS. The active RAS can in turn activate RAF, a Ser/Thr kinase. In the next step mitogen-activated protein kinases (MAP kinases) are activated. (Also known as extracellular signal-regulated kinases - ERKs or MAP/ERK kinase (MEK)). As a result of the last step, ERK1 and ERK2 are activated. The phosphorylated forms of ERKs can continue the cascade by triggering activation more interacting kinases which results in altered transcriptional activity in the nucleus.
- Ligand binding to FPR can also induce the activation of CD38, an ectoenzyme of the surface membrane. As a result of activation NAD molecules will enter the cytoplasm. NAD is converted into cyclic ADP ribose (cADPR), a second messenger which interacts with ryanodine receptors (RyR) on the surface of the rough endoplasmic reticulum. The overall result of the process is increased cytoplasmic Ca levels via the direct pathway described above and also via indirect pathways such as opening of Ca channels in the cell membrane. The sustained increase of Ca is required for directed migration of the cells.
See also
References
- ^ Migeotte I, Communi D, Parmentier M (Dec 2006). "Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses". Cytokine & Growth Factor Reviews. 17 (6): 501–19. doi:10.1016/j.cytogfr.2006.09.009. PMID 17084101.
- ^ Ye RD, Boulay F, Wang JM, Dahlgren C, Gerard C, Parmentier M, Serhan CN, Murphy PM (Jun 2009). "International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family". Pharmacological Reviews. 61 (2): 119–61. doi:10.1124/pr.109.001578. PMC 2745437. PMID 19498085.
- Le Y, Murphy PM, Wang JM (Nov 2002). "Formyl-peptide receptors revisited". Trends in Immunology. 23 (11): 541–8. doi:10.1016/S1471-4906(02)02316-5. PMID 12401407.
- Panaro MA, Acquafredda A, Sisto M, Lisi S, Maffione AB, Mitolo V (2006). "Biological role of the N-formyl peptide receptors". Immunopharmacology and Immunotoxicology. 28 (1): 103–27. doi:10.1080/08923970600625975. hdl:11586/115781. PMID 16684671. S2CID 23082578.
- Braun MC, Wang JM, Lahey E, Rabin RL, Kelsall BL (Jun 2001). "Activation of the formyl peptide receptor by the HIV-derived peptide T-20 suppresses interleukin-12 p70 production by human monocytes". Blood. 97 (11): 3531–6. doi:10.1182/blood.V97.11.3531. PMID 11369647.
- Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I (May 2009). "Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors". Nature. 459 (7246): 574–7. Bibcode:2009Natur.459..574R. doi:10.1038/nature08029. PMID 19387439. S2CID 4302009.
- ^ Yuan S, Ghoshdastider U, Trzaskowski B, Latek D, Debinski A, Pulawski W, Wu R, Gerke V, Filipek S (2012). "The role of water in activation mechanism of human N-formyl peptide receptor 1 (FPR1) based on molecular dynamics simulations". PLOS ONE. 7 (11): e47114. Bibcode:2012PLoSO...747114Y. doi:10.1371/journal.pone.0047114. PMC 3506623. PMID 23189124.
- Lala A, Gwinn M, De Nardin E (Sep 1999). "Human formyl peptide receptor function role of conserved and nonconserved charged residues". European Journal of Biochemistry. 264 (2): 495–9. doi:10.1046/j.1432-1327.1999.00647.x. PMID 10491096.
- Partida-Sánchez S, Cockayne DA, Monard S, Jacobson EL, Oppenheimer N, Garvy B, Kusser K, Goodrich S, Howard M, Harmsen A, Randall TD, Lund FE (Nov 2001). "Cyclic ADP-ribose production by CD38 regulates intracellular calcium release, extracellular calcium influx and chemotaxis in neutrophils and is required for bacterial clearance in vivo". Nature Medicine. 7 (11): 1209–16. doi:10.1038/nm1101-1209. PMID 11689885. S2CID 13239085.
External links
- "Formylpeptide Receptors". IUPHAR Database of Receptors and Ion Channels. International Union of Basic and Clinical Pharmacology. Archived from the original on 2015-06-19. Retrieved 2007-11-02.
- Formyl+peptide+receptor at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
| Innate immune system: Pattern recognition receptors | |
|---|---|
| Membrane-bound_PRRs | |
| Cytoplasmic_PRRs | |
| Secreted_PRRs | |
| Other/ungrouped | |