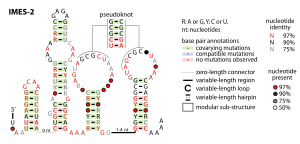
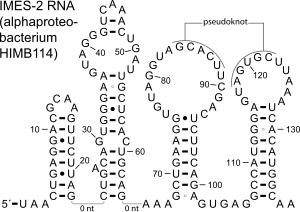
The IMES-2 RNA motif is a conserved RNA structure that was identified by a study based on metagenomics and bioinformatics, and the underlying RNA sequences were identified independently by a similar earlier study. These RNAs are present in environmental sequences, and when discovered were not known to be present in any cultivated species. However, an IMES-2 RNA has been detected in alphaproteobacterium HIMB114, which is classified in the SAR11 clade of marine bacteria. This finding fits with earlier predictions that species that use IMES-2 RNAs are most closely related to alphaproteobacteria. IMES-2 RNAs are exceptionally abundant, as twice as many IMES-2 RNAs were found as ribosomes in RNAs sampled from the Pacific Ocean. Only two bacterial RNAs are known (6S RNA and transfer RNA) to be more highly transcribed than ribosomes.
The IMES-2 RNA secondary structure contains four stem-loop structures and one pseudoknot.
References
- ^ Weinberg Z, Perreault J, Meyer MM, Breaker RR (December 2009). "Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis". Nature. 462 (7273): 656–659. Bibcode:2009Natur.462..656W. doi:10.1038/nature08586. PMC 4140389. PMID 19956260.
- ^ Shi Y, Tyson GW, DeLong EF (May 2009). "Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column". Nature. 459 (7244): 266–269. Bibcode:2009Natur.459..266S. doi:10.1038/nature08055. PMID 19444216. S2CID 4340144.
- Frias-Lopez J, Shi Y, Tyson GW, et al. (March 2008). "Microbial community gene expression in ocean surface waters". Proc. Natl. Acad. Sci. U.S.A. 105 (10): 3805–3810. doi:10.1073/pnas.0708897105. PMC 2268829. PMID 18316740.