| Jaagsiekte sheep retrovirus | |
|---|---|

| |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Pararnavirae |
| Phylum: | Artverviricota |
| Class: | Revtraviricetes |
| Order: | Ortervirales |
| Family: | Retroviridae |
| Genus: | Betaretrovirus |
| Species: | Jaagsiekte sheep retrovirus |
Jaagsiekte sheep retrovirus (JSRV) is a betaretrovirus which is the causative agent of a contagious lung cancer in sheep, called ovine pulmonary adenocarcinoma.
Natural history
JSRV is the virus that is the cause of the contagious lung tumors in sheep called ovine pulmonary adenocarcinoma (OPA). The disease has also been called "jaagsiekte", after the Afrikaans words for "chase" (jaag) and "sickness" (siekte), to describe the respiratory distress observed in an animal out of breath from being chased, indicating the breathing difficulty experienced by infected sheep. Transmission of virus is through aerosol spread between sheep.
The exogenous infectious form of JSRV has an endogenous counterpart which is present in the genomes of all sheep and goats. The sheep genome has around 27 copies of endogenous retroviruses (enJSRVs) that are closely related to JSRV. Endogenous JSRV has several roles in the evolution of the domestic sheep as they are able to block the JSRV replication cycle and play a critical role in sheep conceptus development and placental morphogenesis.
Although OPA resembles human lung cancer, human lung cancer is not known to be caused by betaretroviruses. Even though a possibility of a viral cause has been eliminated in bronchoalveolar cancer, understanding the molecular mechanisms leading to the transformation of lung epithelia by JSRV may be of interest in the context of therapeutic approaches in human lung cancers in general and bronchoalveolar adenocarcinoma (BAC) in particular.
Classification
JSRV belongs to the family Retroviridae, to the subfamily Orthoretrovirinae and the genus Betaretrovirus.
Pathogenesis
JSRV is transmitted by the respiratory route and may also infect lymphocytes and myeloid cells, in addition to the lung epithelia. Expression of the JSRV Envelope protein activates signalling cascades that promote cellular proliferation and malignant transformation of the cells. Initially, the tumour cells grow along the alveolar walls in a pattern reminiscent of human BAC, but subsequently become more invasive and metastasize to the local lymph nodes. Larger tumours may be necrotic and fibromatous at their centre. As the tumour grows, fluid production in the lung increases and this is likely to promote virus spread to other sheep. Only when the tumour reaches a size large enough to compromise lung function, do clinical signs appear. Critically, the majority of infected animals in endemic areas never show outward signs of infection, but they may be shedding virus, thus promoting inadvertent introduction of the disease into previously unaffected flocks and new geographical areas.
Genome structure
The genome of the exogenous virus is 7462 bases and has the classical "gag", "pol", "env" genome arrangement and is flanked by a long terminal repeat (LTR) on each end. There are 4 genes that encode the viral structural proteins. They are "gag" encoding the structural internal virion proteins comprising "matrix" (MA), "capsid" (CA) and "nucleocapsid"(NC); "pro", which encodes an aspartic protease (PR); "pol", which encodes the" RT" and "integrase"(IN) enzymes; and "env", which encodes the "surface" (SU) and "transmembrane" (TM) envelope glycoproteins. The viral proteins are synthesized initially as large precursors and are later processed into the mature proteins by proteolytic cleavage. An additional open reading frame (ORF) was observed in the viral genome and has been called orfX and its function is undefined.
Replication cycle
The initial attachment of JSRV to its target cell is mediated through the binding of the SU subunit of the Env glycoprotein to a specific cell surface receptor molecule,"Hyal2". The entry of the JSRV core into the cytoplasm activates reverse transcription, during which the single-stranded RNA genome is converted into a double-stranded DNA form and gets integrated as a provirus into the host. Following integration, expression of JSRV RNA from the viral promoter in the LTR is controlled by the host transcriptional machine. Following transcription and translation of the viral genome, the new progeny virus gets assembled at the plasma membrane and bud off from the host cell acquiring a lipid envelope and their "env" glycoproteins. Following release from the cell, the "Gag"-"Pro"-"Pol" polyproteins are cleaved into their mature forms by protease. This step maturation is essential for the formation of infectious particles.
Receptor and entry
The cellular receptor for JSRV is hyaluronidase 2 (Hyal2), a glycophosphatidylinol(GPI)-anchored protein belonging to the hyaluronidase family. Generally, oncogenic retroviruses cause transformation of host cells mostly by insertional activation of a host protooncogene into an oncogene. But JSRV is different in this aspect since its envelope glycoprotein ("env") by itself is an oncogene and this single protein was shown to be necessary and sufficient to induce lung tumors in sheep. Unlike the majority of retroviruses, JSRV entry into the host cell is pH-dependent. Thus oncogenic JSRV has borrowed features of both pH-dependent and pH-independent viruses for entry which involves both the receptor binding and a low pH for fusion transformation of host cells.
Host immune response
An important feature of JSRV infection is the absence of any specific immune response from the host. A likely explanation is that the sheep are immunologically tolerant to JSRV antigens due to the expression of closely related endogenous JSRV proteins in the fetal thymus during T lymphocyte development and any JSRV-reactive T cells should be recognized as ‘anti-self’ and selectively removed. Another hypothesis is that tumor cells downregulate their major histocompatibility class-I expression, possibly being the reason for the absence of any virus-specific cytotoxic T cell response (CTL).
Endogenous jaagsiekte sheep retrovirus
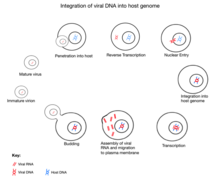
During evolution, the sheep genome incorporated parts of the Jaagksiete sheep retrovirus, now known as endogenous Jaagsiekte sheep retrovirus (enJSRV). There are 27 known copies of enJSRV in the sheep genome, of which five show intact sequences found in all retroviruses. These seem to have been adopted by the sheep genome as enJSRV aids placental development and provides protection against similar retroviruses. In vitro assays have found that enJSRV does this by blocking various stages of the viral replication cycle. An example of this protection is seen in ovine endometrial epithelium where the high expression of enJSVR prevents exogenous JSVR from entering the cells via blocking the common receptor to both, HYAL2. However, Jaagsiekte virus can sometimes mutate to overcome this protection, and there is evidence of this having occurred in the last 200 years. There is also indication that the endogenization of Jaagsiekte virus is still occurring today.
enJSRV mechanism in reproduction

In sheep, enJSRVs are highly expressed in the epithelia lining different reproductive tissues, including the vagina, uterus and oviduct. The RNA of enJSRVs is first detected in the conceptus on day 12. Experiments have found that the enJSRV envelope regulates trophoblast growth and differentiation within the peri-implantation conceptus. It was discovered that enJSRVs are expressed in the trophectoderm cells of the placenta. Their expression coincides with the key events of conceptus elongation and onset of trophoblast giant binucleate cells (BNC) differentiation. Furthermore, it was observed that an injection of morpholinos (an enJSRV envelope production inhibitor) into the uteri of pregnant sheep on day 8 of pregnancy resulted in reduced conceptus elongation and inhibition of trophoblast giant BNC differentiation. Elongation of the sheep conceptus is an essential process as it results in the production of interferon tau (IFNT) which is a pregnancy recognition signal required for conceptus survival. This stimulates both the corpus luteum to continue to secrete progesterone and the onset of implantation. Following the injection of morpholinos, it was observed that pregnancy loss occurred 12 days later. This work supports the hypothesis that enJSRVs are crucial in sheep reproduction and placental morphogenesis.
HYAL2
Hyaluronidase 2 (HYAL2) serves as a cell-surface receptor for both the exogenous and endogenous JSRV envelope (env). HYAL2 mRNA can be detected in the BNCs and multinucleated syncytia of sheep placentomes during pregnancy, but not in the trophectoderm cells or any cells of the endometrium. In situ hybridization analysis revealed that HYAL2 mRNA was only detected in the binucleate cells and multi-nucleated syncytial plaques. It is hypothesised that enJSRV interactions with HYAL2 are vital for placental growth and differentiation. Whilst the cellular and molecular mechanism are still unclear, it is apparent it has a role in protecting the uterus against viral infection and placental morphogenesis.
The co-expression of the enJSRV envelope and HYAL2 in the same cell types supports the hypothesis that HYAL2 binds to enJSRVs env on the binucleate cells and promotes their fusion into multi-nucleated syncytia.
Comparative physiology in humans and mice
Of interest for comparative physiology is that the presence of enJSRV envelope protein expression in the developing sheep placenta is very similar to that observed for syncytin in humans and the mouse. During the formation of the human placenta syncytiotrophoblast, by fusion of mononuclear cytotrophoblasts, human syncytins are specifically expressed. The syncytins are fusogenic when expressed in vitro, supporting the hypothesis that they are involved in placental morphogenesis. These observations support the theory that an ancient retroviral infection had important consequences for mammalian evolution. The involvement of the betaretrovirus enJSRV in the sheep conceptus trophoblasts further argues for its involvement in sheep placentation.
Future directions and summary
Research surrounding endogenous retroviruses supports the idea that they may play critical roles in conceptus growth, placental differentiation and cell fusion in mammals. The morphological aspects of binucleate cell differentiation in ruminants such as sheep are well characterised, but the mechanisms are not well defined - though evidence shows that enJSRV RNA and HYAL2 mRNA are co-expressed in the binucleate cell and multinucleated syncytiotrophoblasts throughout gestation.
See also
References
- ^ York DF, Querat G (2003). "A History of Ovine Pulmonary Adenocarcinoma (Jaagsiekte) and Experiments Leading to the Deduction of the JSRV Nucleotide Sequence". Jaagsiekte Sheep Retrovirus and Lung Cancer. Current Topics in Microbiology and Immunology. Vol. 275. pp. 1–23. doi:10.1007/978-3-642-55638-8_1. ISBN 978-3-642-62897-9. PMID 12596893.
- ^ Arnaud F, Varela M, Spencer TE, Palmarini M (November 2008). "Coevolution of endogenous betaretroviruses of sheep and their host". Cellular and Molecular Life Sciences. 65 (21): 3422–32. doi:10.1007/s00018-008-8500-9. PMC 4207369. PMID 18818869.
- Hopwood P, Wallace WA, Cousens C, Dewar P, Muldoon M, Norval M, Griffiths DJ (November 2010). "Absence of markers of betaretrovirus infection in human pulmonary adenocarcinoma". Human Pathology. 41 (11): 1631–40. doi:10.1016/j.humpath.2010.05.013. PMID 20825971.
- Leroux C, Girard N, Cottin V, Greenland T, Mornex JF, Archer F (2007). "Jaagsiekte Sheep Retrovirus (JSRV): from virus to lung cancer in sheep". Veterinary Research. 38 (2): 211–28. doi:10.1051/vetres:2006060. PMID 17257570.
- ^ Griffiths DJ, Martineau HM, Cousens C (May 2010). "Pathology and pathogenesis of ovine pulmonary adenocarcinoma". Journal of Comparative Pathology. 142 (4): 260–83. doi:10.1016/j.jcpa.2009.12.013. PMID 20163805.
- Caporale M, Cousens C, Centorame P, Pinoni C, De las Heras M, Palmarini M (August 2006). "Expression of the jaagsiekte sheep retrovirus envelope glycoprotein is sufficient to induce lung tumors in sheep". Journal of Virology. 80 (16): 8030–7. doi:10.1128/JVI.00474-06. PMC 1563803. PMID 16873259.
- Côté M, Zheng YM, Liu SL (November 2009). "Receptor binding and low pH coactivate oncogenic retrovirus envelope-mediated fusion". Journal of Virology. 83 (22): 11447–55. doi:10.1128/JVI.00748-09. PMC 2772678. PMID 19726505.
- ^ Armezzani A, Varela M, Spencer TE, Palmarini M, Arnaud F (December 2014). ""Ménage à Trois": the evolutionary interplay between JSRV, enJSRVs and domestic sheep". Viruses. 6 (12): 4926–45. doi:10.3390/v6124926. PMC 4276937. PMID 25502326.
- ^ Varela M, Spencer TE, Palmarini M, Arnaud F (October 2009). "Friendly viruses: the special relationship between endogenous retroviruses and their host". Annals of the New York Academy of Sciences. 1178: 157–72. doi:10.1111/j.1749-6632.2009.05002.x. PMC 4199234. PMID 19845636.
- ^ Dunlap KA, Palmarini M, Adelson DL, Spencer TE (August 2005). "Sheep endogenous betaretroviruses (enJSRVs) and the hyaluronidase 2 (HYAL2) receptor in the ovine uterus and conceptus". Biology of Reproduction. 73 (2): 271–9. doi:10.1095/biolreprod.105.039776. PMID 15788753.
- ^ Black SG, Arnaud F, Palmarini M, Spencer TE (October 2010). "Endogenous retroviruses in trophoblast differentiation and placental development". American Journal of Reproductive Immunology. 64 (4): 255–64. doi:10.1111/j.1600-0897.2010.00860.x. PMC 4198168. PMID 20528833.
- ^ Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE (September 2006). "Endogenous retroviruses regulate periimplantation placental growth and differentiation". Proceedings of the National Academy of Sciences of the United States of America. 103 (39): 14390–5. Bibcode:2006PNAS..10314390D. doi:10.1073/pnas.0603836103. PMC 1599973. PMID 16980413.
- ^ Brooks K, Burns G, Spencer TE (2014). "Conceptus elongation in ruminants: roles of progesterone, prostaglandin, interferon tau and cortisol". Journal of Animal Science and Biotechnology. 5 (1): 53. doi:10.1186/2049-1891-5-53. PMC 4373033. PMID 25810904.
- Dunlap KA, Palmarini M, Spencer TE (April 2006). "Ovine endogenous betaretroviruses (enJSRVs) and placental morphogenesis". Placenta. 27 (Suppl A): S135-40. doi:10.1016/j.placenta.2005.12.009. PMID 16533524.
Further reading
- Palmarini M, Fan H (2003). "Molecular Biology of Jaagsiekte Sheep Retrovirus". Jaagsiekte Sheep Retrovirus and Lung Cancer. Current Topics in Microbiology and Immunology. Vol. 275. pp. 81–115. doi:10.1007/978-3-642-55638-8_4. ISBN 978-3-642-62897-9. PMID 12596896.
- Sharp JM, DeMartini JC (2003). "Natural History of JSRV in Sheep". Jaagsiekte Sheep Retrovirus and Lung Cancer. Current Topics in Microbiology and Immunology. Vol. 275. pp. 55–79. doi:10.1007/978-3-642-55638-8_3. ISBN 978-3-642-62897-9. PMID 12596895.
| Taxon identifiers | |
|---|---|
| Jaagsiekte sheep retrovirus | |