Methylation specific oligonucleotide microarray, also known as MSO microarray, was developed as a technique to map epigenetic methylation changes in DNA of cancer cells.
The general process starts with modification of DNA with bisulfite, specifically to convert unmethylated cytosine in CpG sites to uracil, while leaving methylated cytosines untouched. The modified DNA region of interest is amplified via PCR and during the process, uracils are converted to thymine. The amplicons are labelled with a fluorescent dye and hybridized to oligonucleotide probes that are fixed to a glass slide. The probes differentially bind to cytosine and thymine residues, which ultimately allows discrimination between methylated and unmethylated CpG sites, respectively.
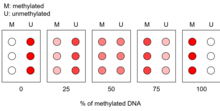
A calibration curve is produced and compared with the microarray results of the amplified DNA samples. This allows a general quantification of the proportion of methylation present in the region of interest.
This microarray technique was developed by Tim Hui-Ming Huang and his laboratory and was officially published in 2002.
Implications for cancer research
| This article needs more reliable medical references for verification or relies too heavily on primary sources. Please review the contents of the article and add the appropriate references if you can. Unsourced or poorly sourced material may be challenged and removed. Find sources: "Methylation specific oligonucleotide microarray" – news · newspapers · books · scholar · JSTOR (November 2019) |  |
Cancer cells often develop atypical methylation patterns, at CpG sites in promoters of tumour suppressor genes. High levels of methylation at a promoter leads to downregulation of the corresponding genes and is characteristic of carcinogenesis. It is one of the most consistent changes observed in early stage tumour cells. Methylation specific oligonucleotide microarray allows for the high resolution and high throughput detection of numerous methylation events on multiple gene promoters. Therefore, this technique can be used to detect aberrant methylation in tumour suppressor promoters at an early stage and has been used in gastric and colon cancers and multiple others. Because it allows one to detect presence of atypical methylations in cancer cells, it can also be used to reveal the major cause behind the malignancy, whether its main contributor is mutations on chromosomes or epigenetic modifications, as well as which tumour suppressor genes' transcription levels are affected. An interesting use of this microarray includes specific classification of cancers based on the methylation patterns alone, such as differentiating between classes of leukemia, suggesting that different classes of cancer show relatively unique methylation patterns. This technique has also been proposed to monitor cancer treatments that involve modifying the methylation patterns in mutant cancer cells.
References
- ^ Gitan RS, Shi H, Chen CM, Yan PS, Huang TH (January 2002). "Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis". Genome Research. 12 (1): 158–64. doi:10.1101/gr.202801. PMC 155260. PMID 11779841.
- ^ Shi H, Maier S, Nimmrich I, Yan PS, Caldwell CW, Olek A, Huang TH (January 2003). "Oligonucleotide-based microarray for DNA methylation analysis: principles and applications". Journal of Cellular Biochemistry. 88 (1): 138–43. doi:10.1002/jcb.10313. PMID 12461783. S2CID 41907903.
- Yan PS, Wei SH, Huang TH (2004). "Methylation-specific oligonucleotide microarray". In Tollefsbol TO (ed.). Epigenetics Protocols. Methods in Molecular Biology. Vol. 287. Humana Press. pp. 251–60. doi:10.1385/1-59259-828-5:251. ISBN 9781592598281. PMID 15273417.
- Hou P, Shen JY, Ji MJ, He NY, Lu ZH (December 2004). "Microarray-based method for detecting methylation changes of p16(Ink4a) gene 5'-CpG islands in gastric carcinomas". World Journal of Gastroenterology. 10 (24): 3553–8. doi:10.3748/wjg.v10.i24.3553. PMC 4611991. PMID 15534905.
- Mund C, Beier V, Bewerunge P, Dahms M, Lyko F, Hoheisel JD (April 2005). "Array-based analysis of genomic DNA methylation patterns of the tumour suppressor gene p16INK4A promoter in colon carcinoma cell lines". Nucleic Acids Research. 33 (8): e73. doi:10.1093/nar/gni072. PMC 1087791. PMID 15860770.
- Yu YP, Paranjpe S, Nelson J, Finkelstein S, Ren B, Kokkinakis D, et al. (February 2005). "High throughput screening of methylation status of genes in prostate cancer using an oligonucleotide methylation array". Carcinogenesis. 26 (2): 471–9. doi:10.1093/carcin/bgh310. PMID 15485992.
- Adorján P, Distler J, Lipscher E, Model F, Müller J, Pelet C, et al. (March 2002). "Tumour class prediction and discovery by microarray-based DNA methylation analysis". Nucleic Acids Research. 30 (5): 21e–21. doi:10.1093/nar/30.5.e21. PMC 101257. PMID 11861926.
External links
- Resources, information and specific protocols for DNA Methylation Analysis
- Software for DNA Methylation Analysis
This genetics article is a stub. You can help Misplaced Pages by expanding it. |
This oncology article is a stub. You can help Misplaced Pages by expanding it. |