| Moco (molybdenum cofactor) riboswitch | |
|---|---|
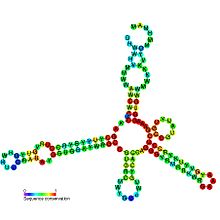 Predicted secondary structure and sequence conservation of MOCO_RNA_motif Predicted secondary structure and sequence conservation of MOCO_RNA_motif | |
| Identifiers | |
| Symbol | MOCO_RNA_motif |
| Rfam | RF01055 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Bacteria |
| SO | SO:0005836 |
| PDB structures | PDBe |
The Moco RNA motif is a conserved RNA structure that is presumed to be a riboswitch that binds molybdenum cofactor or the related tungsten cofactor. Genetic experiments support the hypothesis that the Moco RNA motif corresponds to a genetic control element that responds to changing concentrations of molybdenum or tungsten cofactor. As these cofactors are not available in purified form, in vitro binding assays cannot be performed. However, the genetic data, complex structure of the RNA and the failure to detect a protein involved in the regulation suggest that the Moco RNA motif corresponds to a class of riboswitches.
References
- Weinberg Z, Barrick JE, Yao Z, et al. (2007). "Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline". Nucleic Acids Res. 35 (14): 4809–4819. doi:10.1093/nar/gkm487. PMC 1950547. PMID 17621584.
- ^ Regulski EE, Moy RH, Weinberg Z, et al. (2008). "A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism". Mol. Microbiol. 68 (4): 918–932. doi:10.1111/j.1365-2958.2008.06208.x. PMC 2408646. PMID 18363797.
External links
This molecular or cell biology article is a stub. You can help Misplaced Pages by expanding it. |