| aminoacylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.5.1.14 | ||||||||
| CAS no. | 9012-37-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
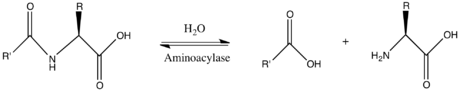
In enzymology, an aminoacylase (EC 3.5.1.14) is an enzyme that catalyzes the chemical reaction
Thus, the two substrates of this enzyme are N-acyl-L-amino acid and H2O, whereas its two products are carboxylate and L-amino acid.
This enzyme belongs to the family of hydrolases, those acting on carbon-nitrogen bonds other than peptide bonds, specifically in linear amides. The systematic name of this enzyme class is N-acyl-L-amino acid amidohydrolase. Other names in common use include dehydropeptidase II, histozyme, hippuricase, benzamidase, acylase I, hippurase, amido acid deacylase, L-aminoacylase, acylase, aminoacylase I, L-amino-acid acylase, alpha-N-acylaminoacid hydrolase, long acyl amidoacylase, and short acyl amidoacylase. This enzyme participates in urea cycle and metabolism of amino groups.
Enzyme structure

As of late 2007, two structures have been solved for this class of enzymes, with PDB accession codes 1Q7L and 1YSJ. These structures also correspond to two known primary amino acid sequences for aminoacylases. The associated papers identify two types of domains comprising aminoacylases: Zinc binding domains - which bind Zn ions - and domains that facilitate dimerization of Zinc binding domains. It is this dimerization that allows catalysis to occur, since aminoacylase's active site lies between its two Zinc binding domains.
Bound Zinc facilitates the binding of the N-acyl-L-amino acid substrate, causing a conformational shift that brings the protein's subunits together around the substrate and allowing catalysis to occur. Aminoacylase 1 exists in a heterotetrameric structure, meaning 2 Zinc binding domains and 2 dimerization domains come together to make aminoacylase 1's quaternary structure.
Enzyme mechanism

Aminoacylase is a metallo-enzyme that needs Zinc (Zn) as a cofactor to function. The Zinc ions inside of aminoacylase are each coordinated to histidine, glutamate, aspartate, and water. The Zinc ion polarizes the water, facilitating its deprotonation by a nearby basic residue. The negatively charged hydroxide ion is nucleophilic and attacks the electrophilic carbonyl carbon of the substrate's acyl group. The exact mechanism after this point is unknown, with one possibility being that the carbonyl then reforms, breaks the amide bond, and forms the two products. At some point in the mechanism, another water molecule enters and coordinates with Zinc, returning the enzyme to its original state.
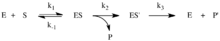
The nucleophilic attack by water is the rate-limiting step of aminoacylase's catalytic mechanism. This nucleophilic attack is reversible while the subsequent steps are fast and irreversible. This reaction sequence is an example of Michaelis–Menten kinetics, allowing one to determine KM, Kcat, Vmax, turnover number, and substrate specificity through classic Michaelis-Menten enzyme experiments. The second and third forward steps cause the formation and release of the reaction's products.
Biological function

Aminoacylases are expressed in the kidney, where they recycle N-acyl-L-amino acids as L-amino acids and aid in urea cycle regulation.
N-acyl-L-amino acids are formed when L-amino acids have their N-terminus covalently bonded to an acyl group. The acyl group provides stability for the amino acid, making it more resistant to degradation. Additionally, N-acyl-L-amino acids cannot be used directly as building blocks for proteins and must first be converted to L-amino acids by aminoacylase. Again, the L-amino acid products can be used for biosynthesis or catabolized energy.
Aminoacylase is involved in the regulation of the urea cycle. N-acetyl-L-glutamate is an allosteric activator of carbamoyl phosphate synthetase, a crucial enzyme that commits NH4 molecules to the urea cycle. The urea cycle gets rid of excess ammonia (NH4) in the body, a process that must be up-regulated during times of increased protein catabolism, as amino acid breakdown produces large amounts of NH4. When amino acid catabolism increases, N-Acetylglutamate synthase is up-regulated, producing more N-acetyl-L-glutamate, which up-regulates carbamoyl phosphate synthetase and allows it to dispose of the excess NH4 from catabolism.
Aminoacylase is up-regulated during times of nutrient deficit or starvation, causing N-acetyl-L-glutamate breakdown, which down-regulates carbamoyl phosphate synthetase and the rest of the urea cycle. This response is evolutionarily advantageous, since a nutrient deficit means there isn't as much NH4 that needs to be disposed of and since the body wants to salvage as many amino acids as it can.
Disease relevance
Aminoacylase 1 deficiency (A1D) is a rare disease caused by an autosomal recessive mutation in the aminoacylase 1 gene (ACY1) on chromosome 3p21. The lack of functional aminoacylase 1 caused by A1D results in a dysfunctional urea cycle, causing an array of neurological disorders including seizures, muscular hypotonia, mental retardation, and impaired psychomotor development. A1D has also been associated with autism . Patients with A1D often start expressing symptoms shortly after birth but seem to recover fully in the next few years.
Aminoacylase 2 deficiency - also known as Canavan's disease - is another rare disease caused by a mutation in the ASPA gene (on chromosome 17) that leads to a deficiency in the enzyme aminoacylase 2. Aminoacylase 2 is known for the fact that it can hydrolyze N-acetylaspartate while aminoacylase 1 cannot.
Industrial relevance
Aminoacylases have been used for the production of L-amino acids in industrial settings since the late 1950s. Since aminoacylases are substrate specific for N-acyl-L-amino acids and not N-acyl-D-amino acids, aminoacylases can be used to reliably take a mixture of these two reactants and only convert the L enantiomers into products - which can then be isolated by solubility from the unreacted N-acyl-D-amino acids. While this process was done in a batch reactor for many years, a faster and less wasteful process was developed in the late 1970s that placed aminoacylases in a column that N-acyl-amino acids were then continuously washed through. This process is still used in industrial settings today to convert N-acyl-amino acids to amino acids in an enantiomerically specific way.
Evolution
Many scientific studies throughout the past half century have used porcine aminoacylase as their model aminoacylase enzyme. The amino acid sequence and primary structure of porcine aminoacylase have been determined. Porcine aminoacylase 1 is composed of two identical heterodimeric subunits each consisting of 406 amino acids, with acetylalanine at the N-terminus of each. Porcine aminoacylase differs from human aminoacylase in structure but replicates its function. It can be inferred from this data that these two enzymes evolved from a common ancestral protein, retaining function but diverging in structure over time.
References
- ^ Lindner HA, Lunin VV, Alary A, Hecker R, Cygler M, Ménard R (November 2003). "Essential roles of zinc ligation and enzyme dimerization for catalysis in the aminoacylase-1/M20 family". The Journal of Biological Chemistry. 278 (45): 44496–504. doi:10.1074/jbc.M304233200. PMID 12933810.
- Fones WS, Lee M (April 1953). "Hydrolysis of N-acyl derivatives of alanine and phenylalanine by acylase I and carboxypeptidase". The Journal of Biological Chemistry. 201 (2): 847–56. doi:10.1016/S0021-9258(18)66242-8. PMID 13061423.
- ^ Lindner HA, Alary A, Wilke M, Sulea T (April 2008). "Probing the acyl-binding pocket of aminoacylase-1". Biochemistry. 47 (14): 4266–75. doi:10.1021/bi702156h. PMID 18341290.
- ^ Mitta M, Ohnogi H, Yamamoto A, Kato I, Sakiyama F, Tsunasawa S (December 1992). "The primary structure of porcine aminoacylase 1 deduced from cDNA sequence". Journal of Biochemistry. 112 (6): 737–42. doi:10.1093/oxfordjournals.jbchem.a123968. PMID 1284246.
- ^ Hernick M, Fierke CA (January 2005). "Zinc hydrolases: the mechanisms of zinc-dependent deacetylases". Archives of Biochemistry and Biophysics. 433 (1): 71–84. doi:10.1016/j.abb.2004.08.006. PMID 15581567.
- ^ Otvös L, Moravcsik E, Mády G (September 1971). "Investigation on the mechanism of acylase-I-catalyzed acylamino acid hydrolysis". Biochemical and Biophysical Research Communications. 44 (5): 1056–64. doi:10.1016/S0006-291X(71)80192-4. PMID 5160398.
- ^ Berg, Jeremy M.; Tymoczko, John L.; Stryer, Lubert (2012). Biochemistry. New York: W. H. Freeman and Company. p. 688. ISBN 978-1-4292-2936-4.
- ^ Sommer A, Christensen E, Schwenger S, et al. (June 2011). "The molecular basis of aminoacylase 1 deficiency" (PDF). Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1812 (6): 685–90. doi:10.1016/j.bbadis.2011.03.005. PMID 21414403.
- Ferri L, Funghini S, Fioravanti A, et al. (October 2013). "Aminoacylase I deficiency due to ACY1 mRNA exon skipping". Clinical Genetics. 86 (4): 367–372. doi:10.1111/cge.12297. PMID 24117009. S2CID 24017306.
- Miller YE, Minna JD, Gazdar AF (June 1989). "Lack of expression of aminoacylase-1 in small cell lung cancer. Evidence for inactivation of genes encoded by chromosome 3p". The Journal of Clinical Investigation. 83 (6): 2120–4. doi:10.1172/JCI114125. PMC 303939. PMID 2542383.
- EntrezGene 95
- Miller YE, Drabkin H, Jones C, Fisher JH (September 1990). "Human aminoacylase-1: cloning, regional assignment to distal chromosome 3p21.1, and identification of a cross-hybridizing sequence on chromosome 18". Genomics. 8 (1): 149–54. doi:10.1016/0888-7543(90)90237-O. PMID 1707030.
- ^ Sass JO, Mohr V, Olbrich H, et al. (March 2006). "Mutations in ACY1, the gene encoding aminoacylase 1, cause a novel inborn error of metabolism". American Journal of Human Genetics. 78 (3): 401–9. doi:10.1086/500563. PMC 1380284. PMID 16465618.
- ^ Sass JO, Olbrich H, Mohr V, et al. (June 2007). "Neurological findings in aminoacylase 1 deficiency". Neurology. 68 (24): 2151–3. doi:10.1212/01.wnl.0000264933.56204.e8. PMID 17562838. S2CID 43376960.
- ^ Van Coster RN, Gerlo EA, Giardina TG, et al. (December 2005). "Aminoacylase I deficiency: a novel inborn error of metabolism". Biochemical and Biophysical Research Communications. 338 (3): 1322–6. doi:10.1016/j.bbrc.2005.10.126. PMID 16274666.
- Tylki-Szymanska A, Gradowska W, Sommer A, et al. (December 2010). "Aminoacylase 1 deficiency associated with autistic behavior". Journal of Inherited Metabolic Disease. 33 Suppl 3: S211–4. doi:10.1007/s10545-010-9089-3. PMID 20480396. S2CID 13374954.
- Xie Q, Guo T, Wang T, Lu J, Zhou HM (November 2003). "Aspartate-induced aminoacylase folding and forming of molten globule". The International Journal of Biochemistry & Cell Biology. 35 (11): 1558–72. doi:10.1016/S1357-2725(03)00131-6. PMID 12824065.
- ^ Sato, Tadashi; Tosa, Tetsuya (2010). "L-Amino Acids Production by Aminoacylase". Encyclopedia of Industrial Biotechnology. pp. 1–20. doi:10.1002/9780470054581.eib497. ISBN 978-0-470-05458-1.
- Birnbaum SM, Levintow L, Kingsley RB, Greenstein JP (January 1952). "Specificity of amino acid acylases". The Journal of Biological Chemistry. 194 (1): 455–70. doi:10.1016/S0021-9258(18)55898-1. PMID 14927637.
- Huang MQ, Zhou HM (1994). "Alkaline unfolding and salt-induced folding of aminoacylase at high pH". Enzyme & Protein. 48 (4): 229–37. doi:10.1159/000474993. PMID 8821711.
- Koreishi M, Asayama F, Imanaka H, et al. (October 2005). "Purification and characterization of a novel aminoacylase from Streptomyces mobaraensis". Bioscience, Biotechnology, and Biochemistry. 69 (10): 1914–22. doi:10.1271/bbb.69.1914. PMID 16244442.
- Mitta M, Kato I, Tsunasawa S (August 1993). "The nucleotide sequence of human aminoacylase-1". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1174 (2): 201–3. doi:10.1016/0167-4781(93)90116-U. PMID 8357837.
| Hydrolases: carbon-nitrogen non-peptide (EC 3.5) | |
|---|---|
| 3.5.1: Linear amides / Amidohydrolases | |
| 3.5.2: Cyclic amides/ Amidohydrolases | |
| 3.5.3: Linear amidines/ Ureohydrolases | |
| 3.5.4: Cyclic amidines/ Aminohydrolases | |
| 3.5.5: Nitriles/ Aminohydrolases | |
| 3.5.99: Other | |
| Enzymes | |
|---|---|
| Activity | |
| Regulation | |
| Classification | |
| Kinetics | |
| Types |
|