| Hepadnaviridae | |
|---|---|

| |
| TEM micrograph showing Hepatitis B virus virions | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Pararnavirae |
| Phylum: | Artverviricota |
| Class: | Revtraviricetes |
| Order: | Blubervirales |
| Family: | Hepadnaviridae |
| Genera | |
Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas (chronic infections), and cirrhosis. It is the sole accepted family in the order Blubervirales.
Taxonomy
The following genera are recognized:
History and discovery
Although liver diseases transmissible among human populations were identified early in the history of medicine, the first known hepatitis with a viral etiological agent was Hepatitis A, in the picornaviridae family. Hepatitis B Virus (HBV) was identified as an infection distinct from Hepatitis A through its contamination of yellow fever vaccine. The vaccine contained human serum as a stabilizing agent which was HBV-infected. HBV was identified as a new DNA virus in the 1960s, followed a couple of decades later by the discovery of the flavivirus hepatitis C. HBV was first identified in the lab as the "Australia agent" by Blumberg and colleagues in the blood of an Aboriginal transfusion patient. This work earned Blumberg the 1976 Nobel Prize in Medicine.
Genome
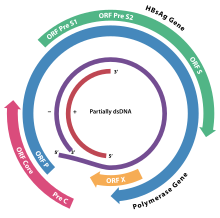
Hepadnaviruses have very small genomes of partially double-stranded, partially single stranded circular DNA (pdsDNA). The genome consists of two strands, a longer negative-sense strand and a shorter and positive-sense strand of variable length. In the virion these strands are arranged such that the two ends of the long strand meet but are not covalently bonded together. The shorter strand overlaps this divide and is connected to the longer strand on either side of the split through a direct repeat (DR) segment that pairs the two strands together. In replication, the viral pdsDNA is converted in the host cell nucleus to covalently-closed-circular DNA (cccDNA) by the viral polymerase.
Replication involves an RNA intermediate, as in viruses belonging to group VII of Baltimore classification. Four main open reading frames are encoded (ORFs) and the virus has four known genes which encode seven proteins: the core capsid protein, the viral polymerase, surface antigens—preS1, preS2, and S, the X protein and HBeAg. The X protein is thought to be non-structural. Its function and significance are poorly understood but it is suspected to be associated with host gene expression modulation.
Viral polymerase
Members of the family Hepadnaviridae encode their own polymerase, rather than co-opting host machinery as some other viruses do. This enzyme is unique among viral polymerases in that it has reverse transcriptase activity to convert RNA into DNA to replicate the genome (the only other human-pathogenic virus family encoding a polymerase with this capability is Retroviridae), RNAse activity (used when the DNA genome is synthesized from pgRNA that was packaged in virions for replication to destroy the RNA template and produce the pdsDNA genome), and DNA-dependent-DNA-polymerase activity (used to create cccDNA from pdsDNA in the first step of the replication cycle).
Envelope proteins
The hepatitis envelope proteins are composed of subunits made from the viral preS1, preS2, and S genes. The L (for "large") envelope protein contains all three subunits. The M (for "medium") protein contains only preS2 and S. The S (for "small") protein contains only S. The genome portions encoding these envelope protein subunits share both the same frame and the same stop codon, generating nested transcripts on a single open reading frame. The pre-S1 is encoded first (closest to the 5' end), followed directly by the pre-S2 and the S. When a transcript is made from the beginning of the pre-S1 region, all three genes are included in the transcript and the L protein is produced. When the transcript starts after the pro-S1 at the beginning of the pre-S2 the final protein contains the pre-S2 and S subunits only and therefore is an M protein. The smallest envelope protein containing just the S subunit is made most because it is encoded closest to the 3' end and comes from the shortest transcript. These envelope proteins can assemble independently of the viral capsid and genome into non-infectious virus-like particles that give the virus a pleomorphic appearance and promote a strong immune response in hosts.
Replication
Hepadnaviruses replicate through an RNA intermediate (which they transcribe back into cDNA using reverse transcriptase). The reverse transcriptase becomes covalently linked to a short 3- or 4-nucleotide primer. Most hepadnaviruses will only replicate in specific hosts, and this makes experiments using in vitro methods very difficult.
The virus binds to specific receptors on cells and the core particle enters the cell cytoplasm. This is then translocated to the nucleus, where the partially double stranded DNA is 'repaired' by the viral polymerase to form a complete circular dsDNA genome (called covalently-closed-circular DNA or cccDNA). The genome then undergoes transcription by the host cell RNA polymerase and the pregenomicRNA (pgRNA) is sent out of the nucleus. The pgRNA is inserted into an assembled viral capsid containing the viral polymerase. Inside this capsid the genome is converted from RNA to pdsDNA through activity of the polymerase as an RNA-dependent-DNA-polymerase and subsequently as an RNAse to eliminate the pgRNA transcript. These new virions either leave the cell to infect others or are immediately dismantled so the new viral genomes can enter the nucleus and magnify the infection. The virions that leave the cell egress through budding.
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Avihepadnavirus | Birds | Hepatocytes | Cell receptor endocytosis | Budding | Nucleus | Cytoplasm | Vertical: parental; sex; blood |
| Orthohepadnavirus | Humans; mammals | Hepatocytes | Cell receptor endocytosis | Budding | Nucleus | Cytoplasm | Vertical: parental; sex; blood |
Structure
Viruses in Hepadnaviridae are enveloped, with spherical geometries, and T=4 symmetry. The diameter is around 42 nm. Genomes are circular, around 3.2kb in length. The genome codes for 7 proteins.
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Avihepadnavirus | Icosahedral | T=4 | Enveloped | Circular | Monopartite |
| Orthohepadnavirus | Icosahedral | T=4 | Enveloped | Circular | Monopartite |
Evolution
Based on the presence of viral genomes in bird DNA it appears that the hepadnaviruses evolved >82 million years ago. Birds may be the original hosts of the Hepadnaviridae with mammals becoming infected after a bird (see host switch).
Endogenous hepatitis B virus genomes have been described in crocodilian, snake and turtle genomes. This suggests that these viruses have infected vertebrates for over 200 million years ago.
Hepadnaviruses have been described in fish and amphibians also. This suggests that this family has co-evolved with the vertebrates.
Phylogenetic trees suggest that the bird viruses originated from those infecting reptiles. Those affecting mammals appear to be more closely related to those found in fish.
Nackednaviridae
A proposed family of viruses – the Nackednaviridae – has been isolated from fish. This family has a similar genomic organisation to that of members of the family Hepadnaviridae. These two families separated over 400 million years ago, suggesting an ancient origin for the family Hepadnaviridae.
Viruses in the family have non-enveloped, isosahedral structure with T=3 symmetry, smaller than typical Hepadnaviridae virions (about 5% of the latter show a T=3 symmetry). The circular, monopartite genome is about 3 kb much like Hepadnaviridae. The envelop protein S is accordingly not present, likely the ancestral state by sequence analysis. Unlike Hepadnaviridae viruses that usually diverge alongside their hosts, viruses in the family jump hosts more frequently. The "type" for this family is African cichlid nackednavirus (ACNDV), formerly African cichlid hepadnavirus (ACHBV), a proposed and not-yet-accepted species.
Cell tropism
Hepadnaviruses, as their "hepa" name implies, infect liver cells and cause hepatitis. This is true not only of the human pathogen Hepatitis B Virus but also the hepadnaviruses that infect other organisms. The "adhesion" step of the dynamic phase—in which an exterior viral protein stably interacts with a host cell protein—determines cell tropism. In the case of HBV the host receptor is human sodium taurocholate receptor (NTCP), a mediator of bile acid uptake, and the virus anti-receptor is the abundant HB-AgS envelope protein.
See also
Notes
- Etymology – portmanteau of hepa (liver: reference to Hepatitis B the primary human member) DNA virus.
References
- "Virus Taxonomy: 2018b Release". International Committee on Taxonomy of Viruses (ICTV). February 2019. Retrieved 14 March 2019.
- Magnius, L; Mason, WS; Taylor, J; Kann, M; Glebe, D; Dény, P; Sureau, C; Norder, H; ICTV Report Consortium (June 2020). "ICTV Virus Taxonomy Profile: Hepadnaviridae". The Journal of General Virology. 101 (6): 571–572. doi:10.1099/jgv.0.001415. PMC 7414443. PMID 32416744.
- ^ "ICTV Report Hepadnaviridae".
- ^ "Viral Zone". ExPASy. Retrieved 15 June 2015.
- "World War II Hepatitis Outbreak Was Biggest in History". AP NEWS. Retrieved 21 June 2023.
- Shin MK, Lee J, Ryu WS (June 2004). "A novel cis-acting element facilitates minus-strand DNA synthesis during reverse transcription of the hepatitis B virus genome". Journal of Virology. 78 (12): 6252–62. doi:10.1128/JVI.78.12.6252-6262.2004. PMC 416504. PMID 15163718.
- Suh A, Brosius J, Schmitz J, Kriegs JO (2013). "The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses". Nature Communications. 4: 1791. Bibcode:2013NatCo...4.1791S. doi:10.1038/ncomms2798. PMID 23653203.
- Suh A, Weber CC, Kehlmaier C, Braun EL, Green RE, Fritz U, Ray DA, Ellegren H (December 2014). "Early Mesozoic coexistence of amniotes and hepadnaviridae". PLOS Genetics. 10 (12): e1004559. doi:10.1371/journal.pgen.1004559. PMC 4263362. PMID 25501991.
- ^ Dill JA, Camus AC, Leary JH, Di Giallonardo F, Holmes EC, Ng TF (September 2016). "Distinct Viral Lineages from Fish and Amphibians Reveal the Complex Evolutionary History of Hepadnaviruses". Journal of Virology. 90 (17): 7920–33. doi:10.1128/JVI.00832-16. PMC 4988138. PMID 27334580.
- ^ Lauber C, Seitz S, Mattei S, Suh A, Beck J, Herstein J, Börold J, Salzburger W, Kaderali L, Briggs JA, Bartenschlager R (September 2017). "Deciphering the Origin and Evolution of Hepatitis B Viruses by Means of a Family of Non-enveloped Fish Viruses". Cell Host & Microbe. 22 (3): 387–399.e6. doi:10.1016/j.chom.2017.07.019. PMC 5604429. PMID 28867387.
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W (November 2012). "Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus". eLife. 1: e00049. doi:10.7554/eLife.00049. PMC 3485615. PMID 23150796.
External links
- ICTV Report: Hepadnaviridae
- Viralzone: Hepadnaviridae
- "Hepadnaviridae". NCBI Taxonomy Browser. 10404.
| Microbiology: Virus | ||
|---|---|---|
| Components |  | |
| Viral life cycle | ||
| Genetics | ||
| By host | ||
| Other | ||
| Taxon identifiers | |
|---|---|
| Hepadnaviridae | |