Phospholipase C, gamma 1, also known as PLCG1 and PLCgamma1, is a protein that in humans involved in cell growth, migration, apoptosis, and proliferation. It is encoded by the PLCG1 gene and is part of the PLC superfamily.
Function
PLCγ1 is a cell growth factor from the PLC superfamily. PLCγ1 is used during cell growth and in cell migration and apoptosis, all of which are vital cell processes that, if disrupted by mutations, can cause cancerous cells to form within the body. Mutations in this protein show an increase in issues in cells regarding regulation of proliferation and their cell signaling. PLCγ1 roles are also involved in neuronal actin growth, calcium signaling, and brain development. It is highly regulated by multiple factors, such as PIK3, AMPK, and FAK. It is part of the PIP3 pathway and leads to and increase in calcium in the cells. In neuronal cells, PLCγ1 is highly involved in actin cytoskeleton organization and synaptic plasticity. The basic PLCγ1 pathway, as scientists currently understand it, is seen below.
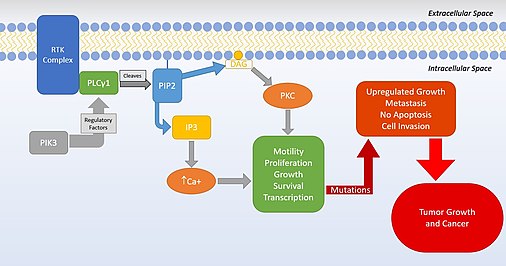
The protein encoded by this gene catalyzes the formation of inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG) from phosphatidylinositol 4,5-bisphosphate. This reaction uses calcium as a cofactor and plays an important role in the intracellular transduction of receptor-mediated tyrosine kinase activators. For example, when activated by SRC, the encoded protein causes the Ras guanine nucleotide exchange factor RASGRP1 to translocate to the Golgi apparatus, where it activates Ras. Also, this protein has been shown to be a major substrate for heparin-binding growth factor 1 (acidic fibroblast growth factor)-activated tyrosine kinase. The receptor protein tyrosine phosphatase PTPmu (PTPRM) is capable of dephosphorylating PLCG1. Two transcript variants encoding different isoforms have been found for this gene.
Common to all PLC isozymes, PLCG1 consists of an N-terminal PH domain, which translocates PLC to the plasma membrane and binds PIP3; four EF hands; an X and Y catalytic region comprising the TIM barrel; and a C-terminal C2 domain. Specific to the PLCG isozymes is a large separation between the X and Y domains consisting of a split PH domain, tandem SH2 domains, and an SH3 domain. The SH2 domains bind phosphorylated tyrosine residues on target proteins via their FLVR sequence motifs, activating the catalytic function of PLCg; and the SH3 domain binds to proline-rich sequences on the target protein.
PLCG1 can be activated by receptor tyrosine kinases (RTKs) and non-receptor tyrosine kinases. For example, when activated, fibroblast growth factor receptor 1 and epidermal growth factor receptor are RTKs that have phosphorylated tyrosines, which provide docking sites for PLCG1 SH2 domains. The activated RTKs phosphoylate PLCG1 at tyrosines located at position 472, 771, 775, 783, and 1254. Non-receptor tyrosine kinases interact with PLCG1 in large complexes at the plasma membrane. For example, in T cells, Lck and Fyn (Src family kinases) phosphorylate immunoreceptor tyrosine-based activation motifs (ITAMs) on the T-cell antigen receptor (TCR). The phosphorylated ITAMs recruit ZAP-70, which phosphorylates tyrosines in LAT and SLP-76. PLCg1 binds to LAT through its n-terminal SH2 domain and to SLP-76 via its SH3 domain.
Has been shown to interact with CISH which negatively regulates it by targeting it for degradation. The deletion of Cish in effector T cells has been shown to augment TCR signaling and subsequent effector cytokine release, proliferation and survival. The adoptive transfer of tumor-specific effector T cells knocked out or knocked down for CISH resulted in a significant increase in functional avidity and long-term tumor immunity. There are no changes in activity or phosphorylation of Cish's purported target, STAT5 in either the presence or absence of Cish.
In vitro studies have shown signs of PLCγ1 having many cell-motility functions, however in vivo have not been able to show a physiological role for PLCγ1. While PLCγ1 is well documented and easily found in the body, clear connections and roles for PLCγ1 have been difficult to find in in vivo studies. Despite this, there is still able to find links between levels of PLCγ1 and cancer patient survivability.
Cancer
Mutations in PLCγ1 can lead to cancer cell proliferations and inhibition can lead to tumor growth. PLCγ1 is involved in cell proliferation, and mutations cause it to be over expressed and help the progression of tumor cells. This aspect of PLCγ1 also helps cancer migration and metastasis away from the original tumor cells. There is also a link between PLCγ1 and PDK, the PDK-PLCγ1 pathway, which is a vital part of cancer cell invasion.
The inhibition of PLCγ1 is linked to a decrease in tumor growth and metastasis. PLCγ1 is acting as a vital part in stopping apoptosis in cells, and thus by inhibiting PLCγ1 the body better allows programmed cell death and avoidance of tumors. The main role found for PLCγ1 is cell growth, and this role in specific is why it is becoming more commonly studied for anti-cancer drugs. Tissue samples from cancer patients the PLCγ1 levels are not elevated, however, regulatory factors for this proteins are lowered and that amplification of PLCγ1 is extremely high. The regulatory proteins that stop PLCγ1 have been turned off by the cell, which means that while there is no increase in the physical protein PLCγ1 there is an increase in how much work it is doing - nothing is stopping it from over working itself. Studies also showed that adding new regulatory to cells in vitro helped reduce previously amplified PLCγ1. This information has encouraged PLCγ1 becoming an anti-cancer drug target despite the issues that come with targeting intermembrane proteins.
Interactions
PLCG1 has been shown to interact with:
- BAG3,
- CD117,
- CD31,
- Cbl gene
- CISH
- Epidermal growth factor receptor,
- Eukaryotic translation elongation factor 1 alpha 1,
- FLT1,
- GAB1,
- GIT1,
- Grb2,
- HER2/neu,
- IRS2,
- ITK,
- KHDRBS1,
- Linker of activated T cells,
- Lymphocyte cytosolic protein 2,
- PDGFRA,
- PLD2,
- RHOA,
- SOS1,
- TUB,
- TrkA,
- TrkB,
- VAV1, and
- Wiskott-Aldrich syndrome protein.
See also
References
- ^ GRCh38: Ensembl release 89: ENSG00000124181 – Ensembl, May 2017
- ^ GRCm38: Ensembl release 89: ENSMUSG00000016933 – Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Bristol A, Hall SM, Kriz RW, Stahl ML, Fan YS, Byers MG, Eddy RL, Shows TB, Knopf JL (1988). "Phospholipase C-148: chromosomal location and deletion mapping of functional domains". Cold Spring Harbor Symposia on Quantitative Biology. 53 (2): 915–20. doi:10.1101/sqb.1988.053.01.105. PMID 3254788.
- Burgess WH, Dionne CA, Kaplow J, Mudd R, Friesel R, Zilberstein A, Schlessinger J, Jaye M (September 1990). "Characterization and cDNA cloning of phospholipase C-gamma, a major substrate for heparin-binding growth factor 1 (acidic fibroblast growth factor)-activated tyrosine kinase". Molecular and Cellular Biology. 10 (9): 4770–7. doi:10.1128/mcb.10.9.4770. PMC 361079. PMID 2167438.
- ^ Koss H, Bunney TD, Behjati S, Katan M (December 2014). "Dysfunction of phospholipase Cγ in immune disorders and cancer" (PDF). Trends in Biochemical Sciences. 39 (12): 603–11. doi:10.1016/j.tibs.2014.09.004. PMID 25456276.
- ^ Lattanzio R, Piantelli M, Falasca M (September 2013). "Role of phospholipase C in cell invasion and metastasis". Advances in Biological Regulation. 53 (3): 309–18. doi:10.1016/j.jbior.2013.07.006. PMID 23925006.
- ^ Jang HJ, Suh PG, Lee YJ, Shin KJ, Cocco L, Chae YC (January 2018). "PLCγ1: Potential arbitrator of cancer progression". Advances in Biological Regulation. 67: 179–189. doi:10.1016/j.jbior.2017.11.003. PMID 29174396.
- ^ Kang DS, Kim IS, Baik JH, Kim D, Cocco L, Suh PG (January 2020). "The function of PLCγ1 in developing mouse mDA system". Advances in Biological Regulation. 75: 100654. doi:10.1016/j.jbior.2019.100654. PMID 31558431. S2CID 203568510.
- Lu X, Fu H, Chen R, Wang Y, Zhan Y, Song G, et al. (2020). "in vitro". International Journal of Biological Sciences. 16 (8): 1427–1440. doi:10.7150/ijbs.42962. PMC 7085223. PMID 32210730.
- Phillips-Mason PJ, Kaur H, Burden-Gulley SM, Craig SE, Brady-Kalnay SM (January 2011). "Identification of phospholipase C gamma1 as a protein tyrosine phosphatase mu substrate that regulates cell migration". Journal of Cellular Biochemistry. 112 (1): 39–48. doi:10.1002/jcb.22710. PMC 3031780. PMID 20506511.
- "Entrez Gene: PLCG1 phospholipase C, gamma 1".
- Singh SM, Murray D (September 2003). "Molecular modeling of the membrane targeting of phospholipase C pleckstrin homology domains". Protein Science. 12 (9): 1934–53. doi:10.1110/ps.0358803. PMC 2323991. PMID 12930993.
- ^ Gresset A, Sondek J, Harden TK (March 2012). "The phospholipase C isozymes and their regulation". Phosphoinositides I: Enzymes of Synthesis and Degradation. Subcellular Biochemistry. Vol. 58. pp. 61–94. doi:10.1007/978-94-007-3012-0_3. ISBN 978-94-007-3011-3. PMC 3638883. PMID 22403074.
- Bae JH, Lew ED, Yuzawa S, Tomé F, Lax I, Schlessinger J (August 2009). "The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site". Cell. 138 (3): 514–24. doi:10.1016/j.cell.2009.05.028. PMC 4764080. PMID 19665973.
- ^ Palmer DC, Guittard GC, Franco Z, Crompton JG, Eil RL, Patel SJ, Ji Y, Van Panhuys N, Klebanoff CA, Sukumar M, Clever D, Chichura A, Roychoudhuri R, Varma R, Wang E, Gattinoni L, Marincola FM, Balagopalan L, Samelson LE, Restifo NP (November 2015). "Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance". The Journal of Experimental Medicine. 212 (12): 2095–113. doi:10.1084/jem.20150304. PMC 4647263. PMID 26527801.
- Kang DS, Kim IS, Baik JH, Kim D, Cocco L, Suh PG (January 2020). "The function of PLCγ1 in developing mouse mDA system". Advances in Biological Regulation. 75: 100654. doi:10.1016/j.jbior.2019.100654. PMID 31558431. S2CID 203568510.
- ^ Lu X, Fu H, Chen R, Wang Y, Zhan Y, Song G, et al. (2020). "in vitro". International Journal of Biological Sciences. 16 (8): 1427–1440. doi:10.7150/ijbs.42962. PMC 7085223. PMID 32210730.
- ^ Jang HJ, Suh PG, Lee YJ, Shin KJ, Cocco L, Chae YC (January 2018). "PLCγ1: Potential arbitrator of cancer progression". Advances in Biological Regulation. 67: 179–189. doi:10.1016/j.jbior.2017.11.003. PMID 29174396.
- ^ Lattanzio R, Piantelli M, Falasca M (September 2013). "Role of phospholipase C in cell invasion and metastasis". Advances in Biological Regulation. 53 (3): 309–18. doi:10.1016/j.jbior.2013.07.006. PMID 23925006.
- Koss H, Bunney TD, Behjati S, Katan M (December 2014). "Dysfunction of phospholipase Cγ in immune disorders and cancer" (PDF). Trends in Biochemical Sciences. 39 (12): 603–11. doi:10.1016/j.tibs.2014.09.004. PMID 25456276.
- Doong H, Price J, Kim YS, Gasbarre C, Probst J, Liotta LA, Blanchette J, Rizzo K, Kohn E (September 2000). "CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70". Oncogene. 19 (38): 4385–95. doi:10.1038/sj.onc.1203797. PMID 10980614.
- van Dijk TB, van Den Akker E, Amelsvoort MP, Mano H, Löwenberg B, von Lindern M (November 2000). "Stem cell factor induces phosphatidylinositol 3'-kinase-dependent Lyn/Tec/Dok-1 complex formation in hematopoietic cells". Blood. 96 (10): 3406–13. doi:10.1182/blood.V96.10.3406. hdl:1765/9530. PMID 11071635.
- Jhun BH, Rivnay B, Price D, Avraham H (April 1995). "The MATK tyrosine kinase interacts in a specific and SH2-dependent manner with c-Kit". The Journal of Biological Chemistry. 270 (16): 9661–6. doi:10.1074/jbc.270.16.9661. PMID 7536744.
- Pumphrey NJ, Taylor V, Freeman S, Douglas MR, Bradfield PF, Young SP, Lord JM, Wakelam MJ, Bird IN, Salmon M, Buckley CD (April 1999). "Differential association of cytoplasmic signalling molecules SHP-1, SHP-2, SHIP and phospholipase C-gamma1 with PECAM-1/CD31". FEBS Letters. 450 (1–2): 77–83. doi:10.1016/s0014-5793(99)00446-9. PMID 10350061. S2CID 31471121.
- ^ Tvorogov D, Carpenter G (July 2002). "EGF-dependent association of phospholipase C-gamma1 with c-Cbl". Experimental Cell Research. 277 (1): 86–94. doi:10.1006/excr.2002.5545. PMID 12061819.
- Graham LJ, Stoica BA, Shapiro M, DeBell KE, Rellahan B, Laborda J, Bonvini E (August 1998). "Sequences surrounding the Src-homology 3 domain of phospholipase Cgamma-1 increase the domain's association with Cbl". Biochemical and Biophysical Research Communications. 249 (2): 537–41. doi:10.1006/bbrc.1998.9177. PMID 9712732.
- Bedrin MS, Abolafia CM, Thompson JF (July 1997). "Cytoskeletal association of epidermal growth factor receptor and associated signaling proteins is regulated by cell density in IEC-6 intestinal cells". Journal of Cellular Physiology. 172 (1): 126–36. doi:10.1002/(SICI)1097-4652(199707)172:1<126::AID-JCP14>3.0.CO;2-A. PMID 9207933. S2CID 24571987.
- Chang JS, Seok H, Kwon TK, Min DS, Ahn BH, Lee YH, Suh JW, Kim JW, Iwashita S, Omori A, Ichinose S, Numata O, Seo JK, Oh YS, Suh PG (May 2002). "Interaction of elongation factor-1alpha and pleckstrin homology domain of phospholipase C-gamma 1 with activating its activity". The Journal of Biological Chemistry. 277 (22): 19697–702. doi:10.1074/jbc.M111206200. PMID 11886851.
- Cunningham SA, Arrate MP, Brock TA, Waxham MN (November 1997). "Interactions of FLT-1 and KDR with phospholipase C gamma: identification of the phosphotyrosine binding sites". Biochemical and Biophysical Research Communications. 240 (3): 635–9. doi:10.1006/bbrc.1997.7719. PMID 9398617.
- Ueno E, Haruta T, Uno T, Usui I, Iwata M, Takano A, Kawahara J, Sasaoka T, Ishibashi O, Kobayashi M (July 2001). "Potential role of Gab1 and phospholipase C-gamma in osmotic shock-induced glucose uptake in 3T3-L1 adipocytes". Hormone and Metabolic Research. 33 (7): 402–6. doi:10.1055/s-2001-16227. PMID 11507676. S2CID 9125865.
- Holgado-Madruga M, Emlet DR, Moscatello DK, Godwin AK, Wong AJ (February 1996). "A Grb2-associated docking protein in EGF- and insulin-receptor signalling". Nature. 379 (6565): 560–4. Bibcode:1996Natur.379..560H. doi:10.1038/379560a0. PMID 8596638. S2CID 4271970.
- Haendeler J, Yin G, Hojo Y, Saito Y, Melaragno M, Yan C, Sharma VK, Heller M, Aebersold R, Berk BC (December 2003). "GIT1 mediates Src-dependent activation of phospholipase Cgamma by angiotensin II and epidermal growth factor". The Journal of Biological Chemistry. 278 (50): 49936–44. doi:10.1074/jbc.M307317200. PMID 14523024.
- Pei Z, Maloney JA, Yang L, Williamson JR (September 1997). "A new function for phospholipase C-gamma1: coupling to the adaptor protein GRB2". Archives of Biochemistry and Biophysics. 345 (1): 103–10. doi:10.1006/abbi.1997.0245. PMID 9281317.
- Nel AE, Gupta S, Lee L, Ledbetter JA, Kanner SB (August 1995). "Ligation of the T-cell antigen receptor (TCR) induces association of hSos1, ZAP-70, phospholipase C-gamma 1, and other phosphoproteins with Grb2 and the zeta-chain of the TCR". The Journal of Biological Chemistry. 270 (31): 18428–36. doi:10.1074/jbc.270.31.18428. PMID 7629168.
- ^ Scholler JK, Perez-Villar JJ, O'Day K, Kanner SB (August 2000). "Engagement of the T lymphocyte antigen receptor regulates association of son-of-sevenless homologues with the SH3 domain of phospholipase Cgamma1". European Journal of Immunology. 30 (8): 2378–87. doi:10.1002/1521-4141(2000)30:8<2378::AID-IMMU2378>3.0.CO;2-E. PMID 10940929.
- Peles E, Levy RB, Or E, Ullrich A, Yarden Y (August 1991). "Oncogenic forms of the neu/HER2 tyrosine kinase are permanently coupled to phospholipase C gamma". The EMBO Journal. 10 (8): 2077–86. doi:10.1002/j.1460-2075.1991.tb07739.x. PMC 452891. PMID 1676673.
- Arteaga CL, Johnson MD, Todderud G, Coffey RJ, Carpenter G, Page DL (December 1991). "Elevated content of the tyrosine kinase substrate phospholipase C-gamma 1 in primary human breast carcinomas". Proceedings of the National Academy of Sciences of the United States of America. 88 (23): 10435–9. Bibcode:1991PNAS...8810435A. doi:10.1073/pnas.88.23.10435. PMC 52943. PMID 1683701.
- Sozzani P, Hasan L, Séguélas MH, Caput D, Ferrara P, Pipy B, Cambon C (March 1998). "IL-13 induces tyrosine phosphorylation of phospholipase C gamma-1 following IRS-2 association in human monocytes: relationship with the inhibitory effect of IL-13 on ROI production". Biochemical and Biophysical Research Communications. 244 (3): 665–70. doi:10.1006/bbrc.1998.8314. PMID 9535722.
- Perez-Villar JJ, Kanner SB (December 1999). "Regulated association between the tyrosine kinase Emt/Itk/Tsk and phospholipase-C gamma 1 in human T lymphocytes". Journal of Immunology. 163 (12): 6435–41. doi:10.4049/jimmunol.163.12.6435. PMID 10586033.
- Hao S, August A (August 2002). "The proline rich region of the Tec homology domain of ITK regulates its activity". FEBS Letters. 525 (1–3): 53–8. doi:10.1016/s0014-5793(02)03066-1. PMID 12163161. S2CID 21541455.
- Oneyama C, Nakano H, Sharma SV (March 2002). "UCS15A, a novel small molecule, SH3 domain-mediated protein-protein interaction blocking drug". Oncogene. 21 (13): 2037–50. doi:10.1038/sj.onc.1205271. PMID 11960376.
- Jabado N, Jauliac S, Pallier A, Bernard F, Fischer A, Hivroz C (September 1998). "Sam68 association with p120GAP in CD4+ T cells is dependent on CD4 molecule expression". Journal of Immunology. 161 (6): 2798–803. doi:10.4049/jimmunol.161.6.2798. PMID 9743338.
- Shen Z, Batzer A, Koehler JA, Polakis P, Schlessinger J, Lydon NB, Moran MF (August 1999). "Evidence for SH3 domain directed binding and phosphorylation of Sam68 by Src". Oncogene. 18 (33): 4647–53. doi:10.1038/sj.onc.1203079. PMID 10467411.
- Zhang W, Trible RP, Samelson LE (August 1998). "LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation". Immunity. 9 (2): 239–46. doi:10.1016/s1074-7613(00)80606-8. PMID 9729044.
- Paz PE, Wang S, Clarke H, Lu X, Stokoe D, Abo A (June 2001). "Mapping the Zap-70 phosphorylation sites on LAT (linker for activation of T cells) required for recruitment and activation of signalling proteins in T cells". The Biochemical Journal. 356 (Pt 2): 461–71. doi:10.1042/0264-6021:3560461. PMC 1221857. PMID 11368773.
- Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE (January 1998). "LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation". Cell. 92 (1): 83–92. doi:10.1016/S0092-8674(00)80901-0. PMID 9489702. S2CID 1806525.
- Yablonski D, Kadlecek T, Weiss A (July 2001). "Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT". Molecular and Cellular Biology. 21 (13): 4208–18. doi:10.1128/MCB.21.13.4208-4218.2001. PMC 87082. PMID 11390650.
- Eriksson A, Nånberg E, Rönnstrand L, Engström U, Hellman U, Rupp E, Carpenter G, Heldin CH, Claesson-Welsh L (March 1995). "Demonstration of functionally different interactions between phospholipase C-gamma and the two types of platelet-derived growth factor receptors". The Journal of Biological Chemistry. 270 (13): 7773–81. doi:10.1074/jbc.270.13.7773. PMID 7535778.
- Jang IH, Lee S, Park JB, Kim JH, Lee CS, Hur EM, Kim IS, Kim KT, Yagisawa H, Suh PG, Ryu SH (May 2003). "The direct interaction of phospholipase C-gamma 1 with phospholipase D2 is important for epidermal growth factor signaling". The Journal of Biological Chemistry. 278 (20): 18184–90. doi:10.1074/jbc.M208438200. PMID 12646582.
- Thodeti CK, Massoumi R, Bindslev L, Sjölander A (July 2002). "Leukotriene D4 induces association of active RhoA with phospholipase C-gamma1 in intestinal epithelial cells". The Biochemical Journal. 365 (Pt 1): 157–63. doi:10.1042/BJ20020248. PMC 1222665. PMID 12071848.
- Kim MJ, Chang JS, Park SK, Hwang JI, Ryu SH, Suh PG (July 2000). "Direct interaction of SOS1 Ras exchange protein with the SH3 domain of phospholipase C-gamma1". Biochemistry. 39 (29): 8674–82. doi:10.1021/bi992558t. PMID 10913276.
- Kapeller R, Moriarty A, Strauss A, Stubdal H, Theriault K, Siebert E, Chickering T, Morgenstern JP, Tartaglia LA, Lillie J (August 1999). "Tyrosine phosphorylation of tub and its association with Src homology 2 domain-containing proteins implicate tub in intracellular signaling by insulin". The Journal of Biological Chemistry. 274 (35): 24980–6. doi:10.1074/jbc.274.35.24980. PMID 10455176.
- Ohmichi M, Decker SJ, Pang L, Saltiel AR (August 1991). "Nerve growth factor binds to the 140 kd trk proto-oncogene product and stimulates its association with the src homology domain of phospholipase C gamma 1" (PDF). Biochemical and Biophysical Research Communications. 179 (1): 217–23. doi:10.1016/0006-291x(91)91357-i. hdl:2027.42/29169. PMID 1715690.
- Qian X, Riccio A, Zhang Y, Ginty DD (November 1998). "Identification and characterization of novel substrates of Trk receptors in developing neurons". Neuron. 21 (5): 1017–29. doi:10.1016/s0896-6273(00)80620-0. PMID 9856458. S2CID 12354383.
- ^ Meakin SO, MacDonald JI, Gryz EA, Kubu CJ, Verdi JM (April 1999). "The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation". The Journal of Biological Chemistry. 274 (14): 9861–70. doi:10.1074/jbc.274.14.9861. PMID 10092678.
- Koch A, Mancini A, Stefan M, Niedenthal R, Niemann H, Tamura T (March 2000). "Direct interaction of nerve growth factor receptor, TrkA, with non-receptor tyrosine kinase, c-Abl, through the activation loop". FEBS Letters. 469 (1): 72–6. doi:10.1016/s0014-5793(00)01242-4. PMID 10708759. S2CID 28312468.
- Suzuki S, Mizutani M, Suzuki K, Yamada M, Kojima M, Hatanaka H, Koizumi S (June 2002). "Brain-derived neurotrophic factor promotes interaction of the Nck2 adaptor protein with the TrkB tyrosine kinase receptor". Biochemical and Biophysical Research Communications. 294 (5): 1087–92. doi:10.1016/S0006-291X(02)00606-X. PMID 12074588.
- Bertagnolo V, Marchisio M, Volinia S, Caramelli E, Capitani S (December 1998). "Nuclear association of tyrosine-phosphorylated Vav to phospholipase C-gamma1 and phosphoinositide 3-kinase during granulocytic differentiation of HL-60 cells". FEBS Letters. 441 (3): 480–4. doi:10.1016/s0014-5793(98)01593-2. PMID 9891995. S2CID 38371954.
- Banin S, Truong O, Katz DR, Waterfield MD, Brickell PM, Gout I (August 1996). "Wiskott-Aldrich syndrome protein (WASp) is a binding partner for c-Src family protein-tyrosine kinases". Current Biology. 6 (8): 981–8. Bibcode:1996CBio....6..981B. doi:10.1016/s0960-9822(02)00642-5. PMID 8805332. S2CID 162267.
- Finan PM, Soames CJ, Wilson L, Nelson DL, Stewart DM, Truong O, Hsuan JJ, Kellie S (October 1996). "Identification of regions of the Wiskott-Aldrich syndrome protein responsible for association with selected Src homology 3 domains". The Journal of Biological Chemistry. 271 (42): 26291–5. doi:10.1074/jbc.271.42.26291. PMID 8824280.
| PDB gallery | |
|---|---|
|
| Hydrolase: esterases (EC 3.1) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3.1.1: Carboxylic ester hydrolases | |||||||||||||||
| 3.1.2: Thioesterase | |||||||||||||||
| 3.1.3: Phosphatase |
| ||||||||||||||
| 3.1.4: Phosphodiesterase | |||||||||||||||
| 3.1.6: Sulfatase | |||||||||||||||
| Nuclease (includes deoxyribonuclease and ribonuclease) |
| ||||||||||||||
| Enzymes | |
|---|---|
| Activity | |
| Regulation | |
| Classification | |
| Kinetics | |
| Types |
|