| Procollagen-proline dioxygenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Alpha subunits of procollagen-proline dioxygenase. Image shows substrate binding region (orange) and the binding groove of tyrosine residues (yellow) Alpha subunits of procollagen-proline dioxygenase. Image shows substrate binding region (orange) and the binding groove of tyrosine residues (yellow) | |||||||||
| Identifiers | |||||||||
| EC no. | 1.14.11.2 | ||||||||
| CAS no. | 9028-06-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Procollagen-proline dioxygenase, commonly known as prolyl hydroxylase, is a member of the class of enzymes known as alpha-ketoglutarate-dependent hydroxylases. These enzymes catalyze the incorporation of oxygen into organic substrates through a mechanism that requires alpha-Ketoglutaric acid, Fe, and ascorbate. This particular enzyme catalyzes the formation of (2S, 4R)-4-hydroxyproline, a compound that represents the most prevalent post-translational modification in the human proteome.
Enzyme mechanism
Procollagen-proline dioxygenase catalyzes the following reaction:
L-proline + alpha-ketoglutaric acid + O2 → (2S, 4R)-4-hydroxyproline + succinate + CO2
The mechanism for the reaction is similar to that of other dioxygenases, and occurs in two distinct stages: In the first, a highly reactive Fe(IV)=O species is produced. Molecular oxygen is bound end-on in an axial position, producing a dioxygen unit. Nucleophilic attack on C2 generates a tetrahedral intermediate, with loss of the double bond in the dioxygen unit and bonds to iron and the alpha carbon of 2-oxoglutarate. Subsequent elimination of CO2 coincides with the formation of the Fe(IV)=O species. The second stage involves the abstraction of the pro-R hydrogen atom from C-4 of the proline substrate followed by radical combination, which yields hydroxyproline.
As a consequence of the reaction mechanism, one molecule of 2-oxoglutarate is decarboxylated, forming succinate. This succinate is hydrolyzed and replaced with another 2-oxoglutarate after each reaction, and it has been concluded that in the presence of 2-oxoglutarate, enzyme-bound Fe is rapidly converted to Fe, leading to inactivation of the enzyme. Ascorbate is utilized as a cofactor to reduce Fe back to Fe.
Enzyme structure
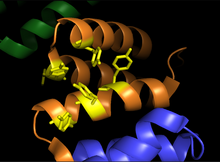
Prolyl hydroxylase is a tetramer with 2 unique subunits. The α subunit is 59 kDa and is responsible for both peptide binding and for catalytic activity. The peptide binding domain spans residues 140-215 of the α subunit, and consists of a concave surface lined with multiple tyrosine residues which interact favorably with the proline-rich substrate. The active site consists of Fe bound to two histidine residues and one aspartate residue, a characteristic shared by most 2-oxoglutarate-dependent dioxygenases. The 55 kDa β subunit is responsible for the enzyme’s localization to and retention in the endoplasmic reticulum. This subunit is identical to the enzyme known as protein disulfide isomerase.
Biological function
Prolyl hydroxylase catalyzes the formation of hydroxyproline. The modification has a significant impact on the stability of collagen, the major connective tissue of the human body. Specifically, hydroxylation increases the melting temperature (Tm) of helical collagen by 16 °C, as compared to unhydroxylated collagen, a difference that allows the protein to be stable at body temperature. Due to the abundance of collagen (about one third of total protein) in humans, and the high occurrence of this modification in collagen, hydroxyproline is quantitatively the most abundant post-translational modification in humans.
The enzyme acts specifically on proline contained within the X-Pro-Gly motif – where Pro is proline. Because of this motif-specific behavior, the enzyme also acts on other proteins that contain this same sequence. Such proteins include C1q, elastins, PrP, Argonaute 2, and conotoxins, among others.
Disease relevance
As prolyl hydroxylase requires ascorbate as a cofactor to function, its absence compromises the enzyme’s activity. The resulting decreased hydroxylation leads to the disease condition known as scurvy. Since stability of collagen is compromised in scurvy patients, symptoms include weakening of blood vessels causing purpura, petechiae, and gingival bleeding.
Hypoxia-inducible factor (HIF) is an evolutionarily conserved transcription factor that allows the cell to respond physiologically to decreases in oxygen. A class of prolyl hydroxylases which act specifically on HIF has been identified; hydroxylation of HIF allows the protein to be targeted for degradation. HIF prolyl-hydroxylase has been targeted by a variety of inhibitors that aim to treat stroke, kidney disease, ischemia, anemia, and other important diseases.
Alternate names
- Protocollagen hydroxylase
- Prolyl hydroxylase
- Prolyl 4-hydroxylase
- Protocollagen prolyl hydroxylase
References
- Smith TG, Talbot NP (April 2010). "Prolyl hydroxylases and therapeutics". Antioxidants & Redox Signaling. 12 (4): 431–3. doi:10.1089/ars.2009.2901. PMID 19761407.
- Hutton JJ, Trappel AL, Udenfriend S (July 1966). "Requirements for alpha-ketoglutarate, ferrous ion and ascorbate by collagen proline hydroxylase". Biochemical and Biophysical Research Communications. 24 (2): 179–84. doi:10.1016/0006-291x(66)90716-9. PMID 5965224.
- ^ Gorres KL, Raines RT (April 2010). "Prolyl 4-hydroxylase". Critical Reviews in Biochemistry and Molecular Biology. 45 (2): 106–24. doi:10.3109/10409231003627991. PMC 2841224. PMID 20199358.
- Fujita Y, Gottlieb A, Peterkofsky B, Udenfriend S, Witkop B (1964). "The Preparation of cis- and trans-4-H3-L-Prolines and Their Use in Studying the Mechanism of Enzymatic Hydroxylation in Chick Embryos". Journal of the American Chemical Society. 86 (21): 4709–4716. doi:10.1021/ja01075a036.
- ^ de Jong L, Albracht SP, Kemp A (June 1982). "Prolyl 4-hydroxylase activity in relation to the oxidation state of enzyme-bound iron. The role of ascorbate in peptidyl proline hydroxylation". Biochimica et Biophysica Acta. 704 (2): 326–32. doi:10.1016/0167-4838(82)90162-5. PMID 6285984.
- De Jong L, Kemp A (May 1984). "Stoicheiometry and kinetics of the prolyl 4-hydroxylase partial reaction". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 787 (1): 105–11. doi:10.1016/0167-4838(84)90113-4. PMID 6326839.
- Berg RA, Prockop DJ (February 1973). "Affinity column purification of protocollagen proline hydroxylase from chick embryos and further characterization of the enzyme". The Journal of Biological Chemistry. 248 (4): 1175–82. doi:10.1016/S0021-9258(19)44278-6. PMID 4346946.
- Helaakoski T, Vuori K, Myllylä R, Kivirikko KI, Pihlajaniemi T (June 1989). "Molecular cloning of the alpha-subunit of human prolyl 4-hydroxylase: the complete cDNA-derived amino acid sequence and evidence for alternative splicing of RNA transcripts". Proceedings of the National Academy of Sciences of the United States of America. 86 (12): 4392–6. Bibcode:1989PNAS...86.4392H. doi:10.1073/pnas.86.12.4392. PMC 287275. PMID 2543975.
- Pekkala M, Hieta R, Bergmann U, Kivirikko KI, Wierenga RK, Myllyharju J (December 2004). "The peptide-substrate-binding domain of collagen prolyl 4-hydroxylases is a tetratricopeptide repeat domain with functional aromatic residues". The Journal of Biological Chemistry. 279 (50): 52255–61. doi:10.1074/jbc.M410007200. PMID 15456751.
- Vuori K, Pihlajaniemi T, Myllylä R, Kivirikko KI (November 1992). "Site-directed mutagenesis of human protein disulphide isomerase: effect on the assembly, activity and endoplasmic reticulum retention of human prolyl 4-hydroxylase in Spodoptera frugiperda insect cells". The EMBO Journal. 11 (11): 4213–7. doi:10.1002/j.1460-2075.1992.tb05515.x. PMC 556932. PMID 1327760.
- Pihlajaniemi T, Helaakoski T, Tasanen K, Myllylä R, Huhtala ML, Koivu J, Kivirikko KI (March 1987). "Molecular cloning of the beta-subunit of human prolyl 4-hydroxylase. This subunit and protein disulphide isomerase are products of the same gene". The EMBO Journal. 6 (3): 643–9. doi:10.1002/j.1460-2075.1987.tb04803.x. PMC 553446. PMID 3034602.
- Shoulders MD, Raines RT (2009). "Collagen structure and stability". Annual Review of Biochemistry. 78: 929–58. doi:10.1146/annurev.biochem.77.032207.120833. PMC 2846778. PMID 19344236.
- Berg RA, Prockop DJ (May 1973). "The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen". Biochemical and Biophysical Research Communications. 52 (1): 115–20. doi:10.1016/0006-291x(73)90961-3. PMID 4712181.
- Gorres, Kelly L.; Raines, Ronald T. (April 2010). "Prolyl 4-hydroxylase". Critical Reviews in Biochemistry and Molecular Biology. 45 (2): 106–124. doi:10.3109/10409231003627991. PMC 2841224. PMID 20199358.
- Müller W, Hanauske-Abel H, Loos M (October 1978). "Biosynthesis of the first component of complement by human and guinea pig peritoneal macrophages: evidence for an independent production of the C1 subunits". Journal of Immunology. 121 (4): 1578–84. doi:10.4049/jimmunol.121.4.1578. PMID 701808. S2CID 36397927.
- Rosenbloom J, Cywinski A (June 1976). "Inhibition of proline hydroxylation does not inhibit secretion of tropoelastin by chick aorta cells". FEBS Letters. 65 (2): 246–50. Bibcode:1976FEBSL..65..246R. doi:10.1016/0014-5793(76)80490-5. PMID 6335. S2CID 34210226.
- Gill AC, Ritchie MA, Hunt LG, Steane SE, Davies KG, Bocking SP, Rhie AG, Bennett AD, Hope J (October 2000). "Post-translational hydroxylation at the N-terminus of the prion protein reveals presence of PPII structure in vivo". The EMBO Journal. 19 (20): 5324–31. doi:10.1093/emboj/19.20.5324. PMC 314005. PMID 11032800.
- Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J, Shi Y (September 2008). "Prolyl 4-hydroxylation regulates Argonaute 2 stability". Nature. 455 (7211): 421–4. Bibcode:2008Natur.455..421Q. doi:10.1038/nature07186. PMC 2661850. PMID 18690212.
- Daly NL, Craik DJ (February 2009). "Structural studies of conotoxins". IUBMB Life. 61 (2): 144–50. doi:10.1002/iub.158. PMID 19165896. S2CID 8678009.
- Bacon NC, Wappner P, O'Rourke JF, Bartlett SM, Shilo B, Pugh CW, Ratcliffe PJ (August 1998). "Regulation of the Drosophila bHLH-PAS protein Sima by hypoxia: functional evidence for homology with mammalian HIF-1 alpha". Biochemical and Biophysical Research Communications. 249 (3): 811–6. doi:10.1006/bbrc.1998.9234. PMID 9731218.
- Smith TG, Robbins PA, Ratcliffe PJ (May 2008). "The human side of hypoxia-inducible factor". British Journal of Haematology. 141 (3): 325–34. doi:10.1111/j.1365-2141.2008.07029.x. PMC 2408651. PMID 18410568.
- ^ Bruick RK, McKnight SL (November 2001). "A conserved family of prolyl-4-hydroxylases that modify HIF". Science. 294 (5545): 1337–40. Bibcode:2001Sci...294.1337B. doi:10.1126/science.1066373. PMID 11598268. S2CID 9695199.
- Karuppagounder SS, Ratan RR (July 2012). "Hypoxia-inducible factor prolyl hydroxylase inhibition: robust new target or another big bust for stroke therapeutics?". Journal of Cerebral Blood Flow and Metabolism. 32 (7): 1347–61. doi:10.1038/jcbfm.2012.28. PMC 3390817. PMID 22415525.
- Warnecke C, Griethe W, Weidemann A, Jürgensen JS, Willam C, Bachmann S, Ivashchenko Y, Wagner I, Frei U, Wiesener M, Eckardt KU (June 2003). "Activation of the hypoxia-inducible factor-pathway and stimulation of angiogenesis by application of prolyl hydroxylase inhibitors". FASEB Journal. 17 (9): 1186–8. doi:10.1096/fj.02-1062fje. PMID 12709400. S2CID 45194703.
- Selvaraju V, Parinandi NL, Adluri RS, Goldman JW, Hussain N, Sanchez JA, Maulik N (June 2014). "Molecular mechanisms of action and therapeutic uses of pharmacological inhibitors of HIF-prolyl 4-hydroxylases for treatment of ischemic diseases". Antioxidants & Redox Signaling. 20 (16): 2631–65. doi:10.1089/ars.2013.5186. PMC 4026215. PMID 23992027.
- Muchnik E, Kaplan J (May 2011). "HIF prolyl hydroxylase inhibitors for anemia". Expert Opinion on Investigational Drugs. 20 (5): 645–56. doi:10.1517/13543784.2011.566861. PMID 21406036. S2CID 21973130.
External links
- Procollagen-proline+dioxygenase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Fe(2+) 2-oxoglutarate dioxygenase domain in PROSITE
| Metabolism: Protein metabolism, synthesis and catabolism enzymes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Essential amino acids are in Capitals | |||||||||
| K→acetyl-CoA |
| ||||||||
| G | |||||||||
| Protein: scleroproteins | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Extracellular matrix |
| ||||||||||||
| Other |
| ||||||||||||
| Oxidoreductases: dioxygenases, including steroid hydroxylases (EC 1.14) | |
|---|---|
| 1.14.11: 2-oxoglutarate | |
| 1.14.13: NADH or NADPH | |
| 1.14.14: reduced flavin or flavoprotein | |
| 1.14.15: reduced iron–sulfur protein | |
| 1.14.16: reduced pteridine (BH4 dependent) | |
| 1.14.17: reduced ascorbate | |
| 1.14.18-19: other | |
| 1.14.99 - miscellaneous | |
| Enzymes | |
|---|---|
| Activity | |
| Regulation | |
| Classification | |
| Kinetics | |
| Types |
|