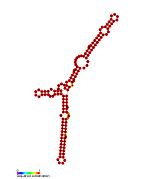
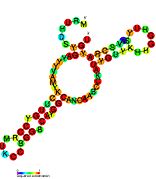
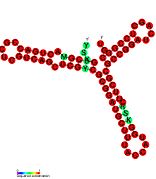
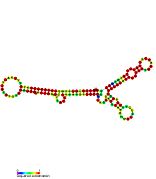
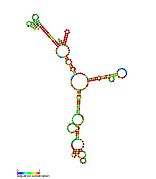
Pseudomonas sRNA are non-coding RNAs (ncRNA) that were predicted by the bioinformatic program SRNApredict2. This program identifies putative sRNAs by searching for co-localization of genetic features commonly associated with sRNA-encoding genes and the expression of the predicted sRNAs was subsequently confirmed by Northern blot analysis. These sRNAs have been shown to be conserved across several pseudomonas species but their function is yet to be determined. Using Tet-Trap genetic approach RNAT genes post-transcriptionally regulated by temperature upshift were identified: ptxS (implicated in virulence) and PA5194.
See also
- Bacillus subtilis sRNA
- Caenorhabditis elegans sRNA
- Mycobacterium tuberculosis sRNA
- Bacteroides thetaiotaomicron sRNA
- NrsZ small RNA
- AsponA antisense RNA
- Repression of heat shock gene expression (ROSE) element
- SrbA sRNA
References
- Livny, J.; Brencic, A.; Lory, S.; Waldor, K. (2006). "Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2" (Free full text). Nucleic Acids Research. 34 (12): 3484–3493. doi:10.1093/nar/gkl453. ISSN 0305-1048. PMC 1524904. PMID 16870723.
- Delvillani, Francesco; Sciandrone, Barbara; Peano, Clelia; Petiti, Luca; Berens, Christian; Georgi, Christiane; Ferrara, Silvia; Bertoni, Giovanni; Pasini, Maria Enrica (December 2014). "Tet-Trap, a genetic approach to the identification of bacterial RNA thermometers: application to Pseudomonas aeruginosa". RNA. 20 (12): 1963–1976. doi:10.1261/rna.044354.114. ISSN 1469-9001. PMC 4238360. PMID 25336583.
| Gallery of Pseudomonas small RNA secondary structure images | |
|---|---|
|