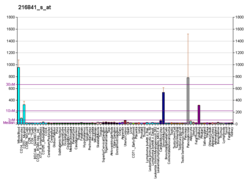
Superoxide dismutase 2, mitochondrial (SOD2), also known as manganese-dependent superoxide dismutase (MnSOD), is an enzyme which in humans is encoded by the SOD2 gene on chromosome 6. A related pseudogene has been identified on chromosome 1. Alternative splicing of this gene results in multiple transcript variants. This gene is a member of the iron/manganese superoxide dismutase family. It encodes a mitochondrial protein that forms a homotetramer and binds one manganese ion per subunit. This protein binds to the superoxide byproducts of oxidative phosphorylation and converts them to hydrogen peroxide and diatomic oxygen. Mutations in this gene have been associated with idiopathic cardiomyopathy (IDC), premature aging, sporadic motor neuron disease, and cancer.
Structure
The SOD2 gene contains five exons interrupted by four introns, an uncharacteristic 5′-proximal promoter that possesses a GC-rich region in place of the TATA or CAAT, and an enhancer in the second intron. The proximal promoter region contains multiple binding sites for transcription factors, including specific-1 (Sp1), activator protein 2 (AP-2), and early growth response 1 (Egr-1). This gene is a mitochondrial member of the iron/manganese superoxide dismutase family. It encodes a mitochondrial matrix protein that forms a homotetramer and binds one manganese ion per subunit. The manganese site forms a trigonal bipyramidal geometry with four ligands from the protein and a fifth solvent ligand. This solvent ligand is a hydroxide believed to serve as the electron acceptor of the enzyme. The active site cavity consists of a network of side chains of several residues associated by hydrogen bonding, extending from the aqueous ligand of the metal. Of note, the highly conserved residue Tyr34 plays a key role in the hydrogen-bonding network, as nitration of this residue inhibits the protein's catalytic ability. This protein also possesses an N-terminal mitochondrial leader sequence which targets it to the mitochondrial matrix, where it converts mitochondrial-generated reactive oxygen species from the respiratory chain to H2. Alternate transcriptional splice variants, encoding different isoforms, have been characterized.
Function
As a member of the iron/manganese superoxide dismutase family, this protein transforms toxic superoxide, a byproduct of the mitochondrial electron transport chain, into hydrogen peroxide and diatomic oxygen. This function allows SOD2 to clear mitochondrial reactive oxygen species (ROS) and, as a result, confer protection against cell death. As a result, this protein plays an antiapoptotic role against oxidative stress, ionizing radiation, and inflammatory cytokines.

Mechanism
SOD2 uses cyclic proton-coupled electron transfer reactions to convert superoxide (O2) into either oxygen (O2) or hydrogen peroxide (H2O2), depending on the oxidation state of the manganese metal and the protonation status of the active site.
Mn + O2 ↔ Mn + O2
Mn + O2 + 2H ↔ Mn + H2O2
The protons of the active site have been directly visualized and revealed that SOD2 utilizes a series of proton transfers among its active site residues per electron transfer step. The findings demonstrate the use of unusual chemistry by the enzyme that include a glutamine that is cyclically deprotonated and protonated and amino acids with pKas that are significantly different from expected values. Low-barrier and short-strong hydrogen bonds are seen contributing to catalysis by promoting proton transfers and stabilizing intermediates in a fashion similar to those of some catalytic Asp-Ser-His triads.
Clinical significance
The SOD2 enzyme is an important constituent in apoptotic signaling and oxidative stress, most notably as part of the mitochondrial death pathway and cardiac myocyte apoptosis signaling. Programmed cell death is a distinct genetic and biochemical pathway essential to metazoans. An intact death pathway is required for successful embryonic development and the maintenance of normal tissue homeostasis. Apoptosis has proven to be tightly interwoven with other essential cell pathways. The identification of critical control points in the cell death pathway has yielded fundamental insights for basic biology, as well as provided rational targets for new therapeutics a normal embryologic processes, or during cell injury (such as ischemia-reperfusion injury during heart attacks and strokes) or during developments and processes in cancer, an apoptotic cell undergoes structural changes including cell shrinkage, plasma membrane blebbing, nuclear condensation, and fragmentation of the DNA and nucleus. This is followed by fragmentation into apoptotic bodies that are quickly removed by phagocytes, thereby preventing an inflammatory response. It is a mode of cell death defined by characteristic morphological, biochemical and molecular changes. It was first described as a "shrinkage necrosis", and then this term was replaced by apoptosis to emphasize its role opposite mitosis in tissue kinetics. In later stages of apoptosis the entire cell becomes fragmented, forming a number of plasma membrane-bounded apoptotic bodies which contain nuclear and or cytoplasmic elements. The ultrastructural appearance of necrosis is quite different, the main features being mitochondrial swelling, plasma membrane breakdown and cellular disintegration. Apoptosis occurs in many physiological and pathological processes. It plays an important role during embryonal development as programmed cell death and accompanies a variety of normal involutional processes in which it serves as a mechanism to remove "unwanted" cells.
Cancer risk
Numerous studies have reported associations between SOD2 polymorphisms and cancer risk, but results have been inconsistent. An updated meta-analysis of such studies revealed that SOD2 polymorphisms are related to the development of non-Hodgkin lymphoma, lung cancer, and colorectal cancer.
Role in oxidative stress
Most notably, SOD2 is pivotal in reactive oxygen species (ROS) release during oxidative stress by ischemia-reperfusion injury, specifically in the myocardium as part of a heart attack (also known as ischemic heart disease). Ischemic heart disease, which results from an occlusion of one of the major coronary arteries, is currently still the leading cause of morbidity and mortality in western society. During ischemia reperfusion, ROS release substantially contribute to the cell damage and death via a direct effect on the cell as well as via apoptotic signals. SOD2 is known to have a capacity to limit the detrimental effects of ROS. As such, SOD2 is important for its cardioprotective effects. In addition, SOD2 has been implicated in cardioprotection against ischemia-reperfusion injury, such as during ischemic preconditioning of the heart. Although a large burst of ROS is known to lead to cell damage, a moderate release of ROS from the mitochondria, which occurs during nonlethal short episodes of ischemia, can play a significant triggering role in the signal transduction pathways of ischemic preconditioning leading to reduction of cell damage. It has even observed that during this release of ROS, SOD2 plays an important role hereby regulating apoptotic signaling and cell death.
Due to its cytoprotective effects, overexpression of SOD2 has been linked to increased invasiveness of tumor metastasis. Its role in controlling ROS levels also involves it in ageing, cancer, and neurodegenerative disease. Mutations in this gene have been associated with idiopathic cardiomyopathy (IDC), sporadic motor neuron disease, and cancer. A common polymorphism associated with greater susceptibility to various pathologies is found in the mitochondrial leader targeting sequence (Val9Ala). Mice lacking Sod2 die shortly after birth, indicating that unchecked levels of superoxide are incompatible with mammalian life. However, mice 50% deficient in Sod2 have a normal lifespan and minimal phenotypic defects but do suffer increased DNA damage and increased incidence of cancer. In Drosophila melanogaster, over-expression of Sod2 has been shown to increase maximum lifespan by 20% in one study, and by as much as 37% in another study.
Yeast studies
In wild-type budding yeast Saccharomyces cerevisiae nuclear DNA fragmentation increased 3-fold during cellular aging, whereas in the absence of SOD2 nuclear DNA fragmentation increased by 5-fold during aging. Production of reactive oxygen species also increased with cellular age, but by a greater amount in SOD2 mutant cells than in wild-type cells. In the fission yeast Schizosaccharomyces pombe, SOD2 deficiency, drastically increased cellular aging and decreased cell viability in the stationary phase of the growth cycle.
Role in invertebrates
SOD2's significant role in oxidative stress management makes it an essential component of the mitochondria. As a result, SOD2 similarly to SOD1 and SOD3 is highly conserved in vertebrates as well as in invertebrates. In the study Multiple measures of functionality exhibit progressive decline in a parallel, stochastic fashion in Drosophilla Sod2 mutants. In SOD2 mutants there was a cascade of deterioration within the organ systems. These deterioration were not linear in that one organ's system would fail then the other, rather on the contrary the deterioration were parallel, meaning that various systems would be affected at any given time. The build up of ROS's in the flies did play a substantial role in affecting the organ system s of the flies in such a way, that though not all observed flies suffered permanent damage, the damages that were observed were like those associated with old age in mature fruit flies. The tissues that are affected in light of defective SOD2 in invertebrates are the muscles, heart, and brain. ROS's effect on these tissue results in not only loss of cellular function in most cases, but a substantial loss in longevity. Though SOD2's role in oxidative stress management is one that has been accepted for both vertebrates and invertebrates, its necessity has been questioned by a study that was conducted on Caenorhabditis elegans (C. elegans). The correlation between the lack of defective SOD2 and loss of longevity and function is generally understood, however it was discovered that the removal of some of the five members of the SOD family including SOD2 resulted in the increase in longevity in mutant C. elegans compared to the wild type.
Animal studies
When animals are exercised at a relatively high work rate, exercise training promotes an increase in myocardial MnSOD activity. Increased MnSOD activity is required to achieve optimal training-induced protection against both ischemia/reperfusion(IR)-induced cardiac arrhythmias and infarction Using an antisense oligonucleotide against MnSOD to prevent ExTr-induced increases in myocardial MnSOD activity, it was demonstrated that an increase in myocardial MnSOD activity is required to provide training-induced protection against IR-induced myocardial infarction. Using a MnSOD gene silencing approach, reported that prevention of the ExTr-induced increase in myocardial MnSOD resulted in a loss of training-induced protection against IR-mediated arrhythmias.
In a mouse model, mitochondrial oxidative stress caused by SOD2 deficiency promoted cellular senescence and aging phenotypes in the skin including an increase in DNA double-strand breaks (see DNA damage theory of aging). Loss of epidermal SOD2 in mice induced cellular senescence, which irreversibly arrested proliferation of a fraction of keratinocytes. In older mice SOD2 deficiency delayed wound closure and reduced epidermal thickness.
Mutant mice with a connective tissue specific lack of SOD2 had a reduced lifespan and a premature onset of aging-related phenotypes such as weight loss, skin atrophy, kyphosis (curvature of the spine), osteoporosis, and muscle degeneration.
SOD2 over-expression was found to extend lifespan in mice.
Interactions
The SOD2 gene has been shown to bind:
The SOD2 protein has been shown to interact with HIV-1 Tat and HIV-1 Vif.
References
- ^ GRCh38: Ensembl release 89: ENSG00000112096 – Ensembl, May 2017
- ^ GRCm38: Ensembl release 89: ENSMUSG00000006818 – Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: SOD2 superoxide dismutase 2, mitochondrial".
- ^ Becuwe P, Ennen M, Klotz R, Barbieux C, Grandemange S (Dec 2014). "Manganese superoxide dismutase in breast cancer: from molecular mechanisms of gene regulation to biological and clinical significance". Free Radical Biology & Medicine. 77: 139–151. doi:10.1016/j.freeradbiomed.2014.08.026. PMID 25224035.
- ^ Pias EK, Ekshyyan OY, Rhoads CA, Fuseler J, Harrison L, Aw TY (Apr 2003). "Differential effects of superoxide dismutase isoform expression on hydroperoxide-induced apoptosis in PC-12 cells". The Journal of Biological Chemistry. 278 (15): 13294–301. doi:10.1074/jbc.M208670200. PMID 12551919.
- ^ Perry JJ, Hearn AS, Cabelli DE, Nick HS, Tainer JA, Silverman DN (Apr 2009). "Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis". Biochemistry. 48 (15): 3417–24. doi:10.1021/bi8023288. PMC 2756076. PMID 19265433.
- ^ Azadmanesh J, Lutz WE, Coates L, Weiss KL, Borgstahl GE (April 2021). "Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase". Nature Communications. 12 (1): 2079. Bibcode:2021NatCo..12.2079A. doi:10.1038/s41467-021-22290-1. PMC 8024262. PMID 33824320.
- Agback P, Agback T (July 2018). "Direct evidence of a low barrier hydrogen bond in the catalytic triad of a Serine protease". Scientific Reports. 8 (1): 10078. Bibcode:2018NatSR...810078A. doi:10.1038/s41598-018-28441-7. PMC 6031666. PMID 29973622.
- Danial NN, Korsmeyer SJ (January 2004). "Cell death: critical control points". Cell. 116 (2): 205–19. doi:10.1016/s0092-8674(04)00046-7. PMID 14744432. S2CID 10764012.
- Kerr JF, Wyllie AH, Currie AR (Aug 1972). "Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics". British Journal of Cancer. 26 (4): 239–57. doi:10.1038/bjc.1972.33. PMC 2008650. PMID 4561027.
- Kang SW (2015). "Superoxide dismutase 2 gene and cancer risk: evidence from an updated meta-analysis". Int J Clin Exp Med. 8 (9): 14647–55. PMC 4658836. PMID 26628947.
- Murray CJ, Lopez AD (May 1997). "Alternative projections of mortality and disability by cause 1990-2020: Global Burden of Disease Study". Lancet. 349 (9064): 1498–504. doi:10.1016/S0140-6736(96)07492-2. PMID 9167458. S2CID 10556268.
- Braunwald E, Kloner RA (Nov 1985). "Myocardial reperfusion: a double-edged sword?". The Journal of Clinical Investigation. 76 (5): 1713–9. doi:10.1172/JCI112160. PMC 424191. PMID 4056048.
- Maslov LN, Naryzhnaia NV, Podoksenov IuK, Prokudina ES, Gorbunov AS, Zhang I, Peĭ ZhM (Jan 2015). "". Rossiĭskii Fiziologicheskiĭ Zhurnal Imeni I.M. Sechenova / Rossiĭskaia Akademiia Nauk. 101 (1): 3–24. PMID 25868322.
- Liem DA, Honda HM, Zhang J, Woo D, Ping P (Dec 2007). "Past and present course of cardioprotection against ischemia-reperfusion injury". Journal of Applied Physiology. 103 (6): 2129–36. doi:10.1152/japplphysiol.00383.2007. PMID 17673563. S2CID 24815784.
- Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H (Aug 2007). "Trends in oxidative aging theories". Free Radical Biology & Medicine. 43 (4): 477–503. doi:10.1016/j.freeradbiomed.2007.03.034. PMID 17640558.
- Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ (Dec 1995). "Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase". Nature Genetics. 11 (4): 376–81. doi:10.1038/ng1295-376. PMID 7493016. S2CID 10900822.
- ^ Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A (Dec 2003). "Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging". Physiological Genomics. 16 (1): 29–37. doi:10.1152/physiolgenomics.00122.2003. PMID 14679299.
- ^ Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavaré S, Tower J (2007). "Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes". Genome Biology. 8 (12): R262. doi:10.1186/gb-2007-8-12-r262. PMC 2246264. PMID 18067683.
- Sun J, Folk D, Bradley TJ, Tower J (June 2002). "Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster". Genetics. 161 (2): 661–72. doi:10.1093/genetics/161.2.661. PMC 1462135. PMID 12072463.
- Muid KA, Karakaya HÇ, Koc A (February 2014). "Absence of superoxide dismutase activity causes nuclear DNA fragmentation during the aging process". Biochem. Biophys. Res. Commun. 444 (2): 260–3. doi:10.1016/j.bbrc.2014.01.056. hdl:11147/5542. PMID 24462872.
- Ogata T, Senoo T, Kawano S, Ikeda S (January 2016). "Mitochondrial superoxide dismutase deficiency accelerates chronological aging in the fission yeast Schizosaccharomyces pombe". Cell Biol. Int. 40 (1): 100–6. doi:10.1002/cbin.10556. PMID 26507459. S2CID 205563521.
- Piazza N, Hayes M, Martin I, Duttaroy A, Grotewiel M, Wessells R (2009). "Multiple measures of functionality exhibit progressive decline in a parallel, stochastic fashion in Drosophila Sod2 null mutants". Biogerontology. 10 (5): 637–48. doi:10.1007/s10522-008-9210-2. PMC 2800787. PMID 19148770.
- Van Raamsdonk JM, Hekimi S (February 2009). "Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans". PLOS Genetics. 5 (2): e1000361. doi:10.1371/journal.pgen.1000361. PMC 2628729. PMID 19197346.
- Yamashita N, Hoshida S, Otsu K, Asahi M, Kuzuya T, Hori M (1999). "Exercise provides direct biphasic cardioprotection via manganese superoxide dismutase activation". The Journal of Experimental Medicine. 189 (11): 1699–706. doi:10.1084/jem.189.11.1699. PMC 2193084. PMID 10359573.
- Hamilton KL, Quindry JC, French JP, Staib J, Hughes J, Mehta JL, Powers SK (2004). "MnSOD antisense treatment and exercise-induced protection against arrhythmias". Free Radical Biology & Medicine. 37 (9): 1360–8. doi:10.1016/j.freeradbiomed.2004.07.025. PMID 15454275.
- Velarde MC, Flynn JM, Day NU, Melov S, Campisi J (January 2012). "Mitochondrial oxidative stress caused by Sod2 deficiency promotes cellular senescence and aging phenotypes in the skin". Aging (Albany NY). 4 (1): 3–12. doi:10.18632/aging.100423. PMC 3292901. PMID 22278880.
- Velarde MC, Demaria M, Melov S, Campisi J (August 2015). "Pleiotropic age-dependent effects of mitochondrial dysfunction on epidermal stem cells". Proc. Natl. Acad. Sci. U.S.A. 112 (33): 10407–12. Bibcode:2015PNAS..11210407V. doi:10.1073/pnas.1505675112. PMC 4547253. PMID 26240345.
- Treiber N, Maity P, Singh K, Kohn M, Keist AF, Ferchiu F, Sante L, Frese S, Bloch W, Kreppel F, Kochanek S, Sindrilaru A, Iben S, Högel J, Ohnmacht M, Claes LE, Ignatius A, Chung JH, Lee MJ, Kamenisch Y, Berneburg M, Nikolaus T, Braunstein K, Sperfeld AD, Ludolph AC, Briviba K, Wlaschek M, Florin L, Angel P, Scharffetter-Kochanek K (April 2011). "Accelerated aging phenotype in mice with conditional deficiency for mitochondrial superoxide dismutase in the connective tissue". Aging Cell. 10 (2): 239–54. doi:10.1111/j.1474-9726.2010.00658.x. PMID 21108731. S2CID 46458295.
- Hu D, Cao P, Thiels E, Chu CT, Wu GY, Oury TD, Klann E (March 2007). "Hippocampal long-term potentiation, memory, and longevity in mice that overexpress mitochondrial superoxide dismutase". Neurobiol Learn Mem. 87 (3): 372–84. doi:10.1016/j.nlm.2006.10.003. PMC 1847321. PMID 17129739.
- Woollard SM, Bhargavan B, Yu F, Kanmogne GD (Jun 2014). "Differential effects of Tat proteins derived from HIV-1 subtypes B and recombinant CRF02_AG on human brain microvascular endothelial cells: implications for blood-brain barrier dysfunction". Journal of Cerebral Blood Flow and Metabolism. 34 (6): 1047–59. doi:10.1038/jcbfm.2014.54. PMC 4050250. PMID 24667918.
Further reading
- Zelko IN, Mariani TJ, Folz RJ (Aug 2002). "Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression". Free Radical Biology & Medicine. 33 (3): 337–49. doi:10.1016/S0891-5849(02)00905-X. PMID 12126755.
- Faraci FM, Didion SP (Aug 2004). "Vascular protection: superoxide dismutase isoforms in the vessel wall". Arteriosclerosis, Thrombosis, and Vascular Biology. 24 (8): 1367–73. doi:10.1161/01.ATV.0000133604.20182.cf. PMID 15166009.
| PDB gallery | |
|---|---|
|
| Other oxidoreductases (EC 1.15–1.21) | |
|---|---|
| 1.15: Acting on superoxide as acceptor | |
| 1.16: Oxidizing metal ions | |
| 1.17: Acting on CH or CH2 groups | |
| 1.18: Acting on iron–sulfur proteins as donors | |
| 1.19: Acting on reduced flavodoxin as donor | |
| 1.20: Acting on phosphorus or arsenic in donors | |
| 1.21: Acting on X-H and Y-H to form an X-Y bond | |