Trans-acting siRNA (abbreviated "ta-siRNA" or "tasiRNA") are a class of small interfering RNA (siRNA) that repress gene expression through post-transcriptional gene silencing in land plants. Precursor transcripts from TAS loci are polyadenylated and converted to double-stranded RNA, and are then processed into 21-nucleotide-long RNA duplexes with overhangs. These segments are incorporated into an RNA-induced silencing complex (RISC) and direct the sequence-specific cleavage of target mRNA. Ta-siRNAs are classified as siRNA because they arise from double-stranded RNA (dsRNA).
Discovery
ta-siRNA were originally detected in 2004 in the flowering plant Arabidopsis thaliana. Initial descriptions found involvement of the plant protein suppressor of gene silencing 3 (SGS3), and the enzyme RNA-dependant RNA polymerase 6 (RDR6).
Biogenesis
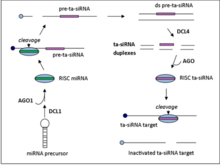
Ta-siRNAs are generated from non-coding transcripts through Argonaute-mediated miRNA-guided cleavage followed by conversion to double stranded RNA by RDR6. The resulting dsRNA is further processed by dicer-like enzyme 4 (DCL4) to produce a phased array of 21-nt siRNAs from positions adjoining the miRNA cleavage site.
There are four families of ta-siRNA-generating loci (TAS genes) in A. thaliana. TAS1, TAS2, and TAS4 families require one miRNA binding site for cleavage to occur while TAS3 requires two binding sites. TAS gene family numbers do not generally indicate orthology, e.g. the moss TAS1 gene family does not share an ancestor gene with the Arabidopsis thaliana TAS1 gene family.
TAS1 and TAS2
TAS1/2 transcripts undergo an initial AGO1 mediated cleavage at the 5’ end that is guided by miR173. RDR6 then converts the transcript into a double strand RNA fragment which then gets processed by DCL4 to generate the 21-nt siRNA with 2 nucleotide 3’ overhangs that target complementary mRNAs in trans.
TAS4
The initial steps for the TAS4 family of ta-siRNA is similar to that of TAS1 and TAS2. The TAS4 family of transcripts first undergo miR828 guided, AGO1 mediated cleavage, followed by dsRNA synthesis and processing by DCL4.
TAS3
In contrast to the single mRNA binding family, TAS3 requires the guide mRNA miR390 bind the transcript at two sites. The transcript is then cleaved at the 3’ binding site only, by AGO7. As is the case for the TAS1, TAS2, and TAS3 families, RDR6 then synthesizes the dsRNA fragment which is further processed by DCL4.
Mechanism
Endogenous ta-siRNAs act via hetero-silencing, which means that the genes they target for cleavage and repression do not have much resemblance to the genes from which the siRNAs derive. This differs from other endogenous siRNAs which are cis-acting and perform auto-silencing, repressing the expression of genes that are the same as or have a lot of resemblance to the genes from which they derive. It was previously thought that only miRNAs exhibited hetero-silencing. Like other siRNAs, the ta-siRNAs are incorporated into RNA-induced silencing complexes (RISCs), where they guide the complex to cleave the target mRNAs in the middle of a single complementary site and repress translation.
A member of the Argonaute protein family is a component of all RNA silencing effector complexes, including the RISCs that catalyze mRNA cleavage. Specifically in arabidopsis, it appears to be AGO7/ZIPPY that plays a role in the ta-siRNA pathway by acting during TAS3-derived ta-siRNA-mediated regulation. AGO7/ZIPPY does not play a role in the mechanisms for TAS1 or TAS2 ta-siRNA biogenesis. ta-siRNAs can be loaded into AGO1 complexes to guide target mRNA cleavage.
Presence in plants
In addition to being present in A. thaliana, evidence of ta-siRNAs has also been found in the moss Physcomitrella patens, maize, Oryza sativa (rice), and other plants. TAS3 trans-acting short-interfering RNA targeting auxin response factors ("tasiR-ARF") is an example of a ta-siRNA that has been shown to be present not only in arabidopsis, but in all of the previous examples. TasiR-ARF is responsible for regulating the signaling molecule auxin. It does this by targeting the mRNA that encodes several of the Auxin Response Factor (ARF) genes for degradation.
References
- ^ Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P (October 2004). "Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs". Mol. Cell. 16 (1): 69–79. doi:10.1016/j.molcel.2004.09.028. PMID 15469823.
- ^ Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (October 2004). "SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis". Genes Dev. 18 (19): 2368–79. doi:10.1101/gad.1231804. PMC 522987. PMID 15466488.
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP (November 2006). "A two-hit trigger for siRNA biogenesis in plants". Cell. 127 (3): 565–77. doi:10.1016/j.cell.2006.09.032. PMID 17081978.
- Axtell, Michael J. (29 April 2013). "Classification and Comparison of Small RNAs from Plants". Annual Review of Plant Biology. 64 (1): 137–159. doi:10.1146/annurev-arplant-050312-120043. PMID 23330790.
- ^ Talmor-Neiman M, Stav R, Klipcan L, Buxdorf K, Baulcombe DC, Arazi T (November 2006). "Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis". Plant J. 48 (4): 511–21. doi:10.1111/j.1365-313X.2006.02895.x. PMID 17076803.
- ^ Allen E, Xie Z, Gustafson AM, Carrington JC (April 2005). "microRNA-directed phasing during trans-acting siRNA biogenesis in plants". Cell. 121 (2): 207–21. doi:10.1016/j.cell.2005.04.004. PMID 15851028.
- ^ Allen E, Howell MD (October 2010). "miRNAs in the biogenesis of trans-acting siRNAs in higher plants". Semin. Cell Dev. Biol. 21 (8): 798–804. doi:10.1016/j.semcdb.2010.03.008. PMID 20359543.
- ^ Tomari Y, Zamore PD (March 2005). "Perspective: machines for RNAi". Genes Dev. 19 (5): 517–29. doi:10.1101/gad.1284105. PMID 15741316.
- ^ Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H (May 2006). "DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7". Curr. Biol. 16 (9): 927–32. doi:10.1016/j.cub.2006.03.035. PMID 16682354.
- Wu L, Mao L, Qi Y (October 2012). "Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation". Plant Physiol. 160 (2): 990–9. doi:10.1104/pp.112.200279. PMC 3461571. PMID 22846193.
- ^ Williams L, Carles CC, Osmont KS, Fletcher JC (July 2005). "A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the arabidopsis ARF2, ARF3, and ARF4 genes". Proc. Natl. Acad. Sci. U.S.A. 102 (27): 9703–8. Bibcode:2005PNAS..102.9703W. doi:10.1073/pnas.0504029102. PMC 1172271. PMID 15980147.
- Heisel SE, Zhang Y, Allen E, Guo L, Reynolds TL, Yang X, Kovalic D, Roberts JK (2008). "Characterization of unique small RNA populations from rice grain". PLOS ONE. 3 (8): e2871. Bibcode:2008PLoSO...3.2871H. doi:10.1371/journal.pone.0002871. PMC 2518513. PMID 18716673.
| Types of RNA | |
|---|---|
| Protein synthesis | |
| RNA processing | |
| Gene regulation | |
| Cis-regulatory elements | |
| Parasites | |
| Other | |
| Types of nucleic acids | |||||||
|---|---|---|---|---|---|---|---|
| Constituents | |||||||
| Ribonucleic acids (coding, non-coding) |
| ||||||
| Deoxyribonucleic acids | |||||||
| Analogues | |||||||
| Cloning vectors | |||||||