The vitamin D receptor (VDR also known as the calcitriol receptor) is a member of the nuclear receptor family of transcription factors. Calcitriol (the active form of vitamin D, 1,25-(OH)2vitamin D3) binds to VDR, which then forms a heterodimer with the retinoid-X receptor. The VDR heterodimer then enters the nucleus and binds to Vitamin D responsive elements (VDRE) in genomic DNA. VDR binding results in expression or transrepression of many specific gene products. VDR is also involved in microRNA-directed post transcriptional mechanisms. In humans, the vitamin D receptor is encoded by the VDR gene located on chromosome 12q13.11.
VDR is expressed in most tissues of the body, and regulates transcription of genes involved in intestinal and renal transport of calcium and other minerals. Glucocorticoids decrease VDR expression. Many types of immune cells also express VDR.
Function
The VDR gene encodes the nuclear hormone receptor for vitamin D. The most potent natural agonist is calcitriol (1,25-dihydroxycholecalciferol) and the vitamin D2 homologue ercalcitriol, 1-alpha,25-dihydroergocalciferol) is also a strong activator. Other forms of vitamin D bind with lower affinity, as does the secondary bile acid lithocholic acid. The receptor belongs to the family of trans-acting transcriptional regulatory factors and shows similarity of sequence to the steroid and thyroid hormone receptors.
Downstream targets of this nuclear hormone receptor include many genes involved in mineral metabolism. The receptor regulates a variety of other metabolic pathways, such as those involved in the immune response and cancer. VDR variants that bolster vitamin-D action and that are directly correlated with AIDS progression rates and VDR association with progression to AIDS follows an additive model. FokI polymorphism is a risk factor for enveloped virus infection as revealed in a meta-analysis. The importance of this gene has also been noted in the natural aging process were 3’UTR haplotypes of the gene showed an association with longevity.
Clinical relevance
Mutations in this gene are associated with type II vitamin D-resistant rickets. A single nucleotide polymorphism in the initiation codon results in an alternate translation start site three codons downstream. Alternative splicing results in multiple transcript variants encoding the same protein. VDR gene variants seem to influence many biological endpoints, including those related to osteoporosis
The vitamin D receptor plays an important role in regulating the hair cycle. Loss of VDR is associated with hair loss in experimental animals. Experimental studies have shown that the unliganded VDR interacts with regulatory regions in cWnt (wnt signaling pathway) and sonic hedgehog target genes and is required for the induction of these pathways during the postnatal hair cycle. These studies have revealed novel actions of the unliganded VDR in regulating the post-morphogenic hair cycle.
Researchers have focused their efforts in elucidating the role of VDR polymorphisms in different diseases and normal phenotypes such as the HIV-1 infection susceptibility and progression or the natural aging process. The most remarkable findings include the report of VDR variants that bolster vitamin-D action and that are directly correlated with AIDS progression rates, that VDR association with progression to AIDS follows an additive model and the role of FokI polymorphism as a risk factor for enveloped virus infection as revealed in a meta-analysis.
Interactions
Vitamin D receptor has been shown to interact with many other factors which will affect transcription activation:
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.
[[File: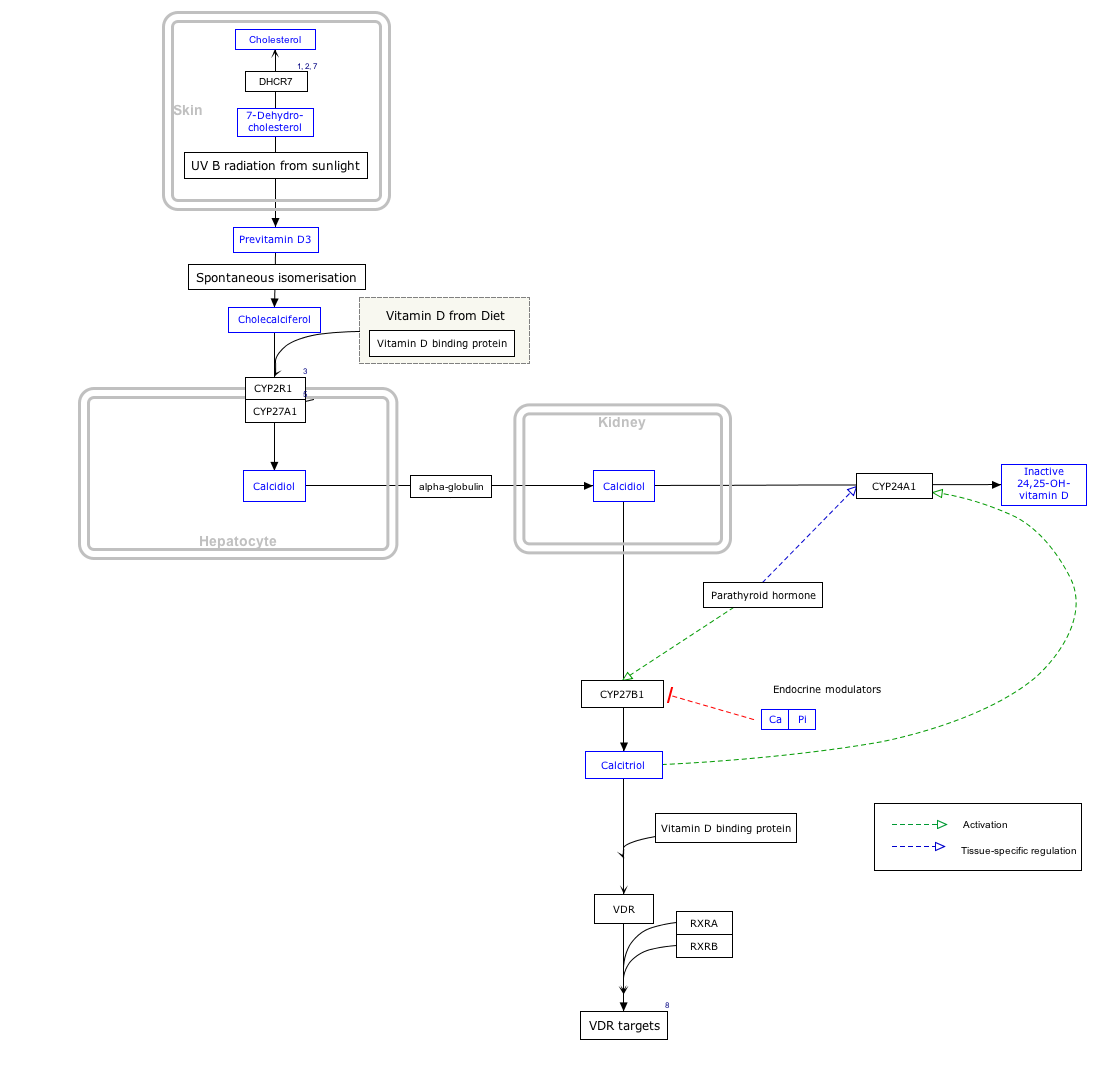

- The interactive pathway map can be edited at WikiPathways: "VitaminDSynthesis_WP1531".
References
- ^ GRCh38: Ensembl release 89: ENSG00000111424 – Ensembl, May 2017
- ^ GRCm38: Ensembl release 89: ENSMUSG00000022479 – Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Moore DD, Kato S, Xie W, Mangelsdorf DJ, Schmidt DR, Xiao R, Kliewer SA (December 2006). "International Union of Pharmacology. LXII. The NR1H and NR1I receptors: constitutive androstane receptor, pregnene X receptor, farnesoid X receptor alpha, farnesoid X receptor beta, liver X receptor alpha, liver X receptor beta, and vitamin D receptor". Pharmacol. Rev. 58 (4): 742–59. doi:10.1124/pr.58.4.6. PMID 17132852. S2CID 85996383.
- Lisse TS, Chun RF, Rieger S, Adams JS, Hewison M (June 2013). "Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts". J Bone Miner Res. 28 (6): 1478–14788. doi:10.1002/jbmr.1882. PMC 3663893. PMID 23362149.
- Szpirer J, Szpirer C, Riviere M, Levan G, Marynen P, Cassiman JJ, Wiese R, DeLuca HF (September 1991). "The Sp1 transcription factor gene (SP1) and the 1,25-dihydroxyvitamin D3 receptor gene (VDR) are colocalized on human chromosome arm 12q and rat chromosome 7". Genomics. 11 (1): 168–73. doi:10.1016/0888-7543(91)90114-T. PMID 1662663.
- ^ Fleet JC, Schoch RD (August 2010). "Molecular Mechanisms for Regulation of Intestinal Calcium Absorption by Vitamin D and Other Factors". Crit Rev Clin Lab Sci. 47 (4): 181–195. doi:10.3109/10408363.2010.536429. PMC 3235806. PMID 21182397.
- ^ Adorini L, Daniel KC, Penna G (2006). "Vitamin D receptor agonists, cancer and the immune system: an intricate relationship". Curr Top Med Chem. 6 (12): 1297–301. doi:10.2174/156802606777864890. PMID 16848743.
- Germain P, Staels B, Dacquet C, Spedding M, Laudet V (December 2006). "Overview of nomenclature of nuclear receptors". Pharmacol. Rev. 58 (4): 685–704. doi:10.1124/pr.58.4.2. PMID 17132848. S2CID 1190488.
- ^ Laplana M, Sánchez-de-la-Torre M, Puig T, Caruz A, Fibla J (July 2014). "Vitamin-D pathway genes and HIV-1 disease progression in injection drug users". Gene. 545 (1): 163–9. doi:10.1016/j.gene.2014.04.035. hdl:10459.1/67999. PMID 24768180.
- ^ Laplana M, Royo L, Fibla J (December 2018). "Vitamin D Receptor polymorphisms and risk of enveloped virus infection: A meta-analysis". Gene. 678: 384–94. doi:10.1016/j.gene.2018.08.017. hdl:10459.1/68000. PMID 30092343. S2CID 51955566.
- Laplana M, Sánchez-de-la-Torre M, Aguiló A, Casado I, Flores M, Sánchez-Pellicer R, Fibla J (April 2010). "Tagging long-lived individuals through vitamin-D receptor (VDR) haplotypes". Biogerontology. 11 (4): 437–46. doi:10.1007/s10522-010-9273-8. hdl:10459.1/67920. PMID 20407924. S2CID 34809120.
- "Entrez Gene: VDR vitamin D (1,25- dihydroxyvitamin D3) receptor".
- Abouzid M, Karazniewicz-Lada M, Glowka F (2018-10-19). "Genetic Determinants of Vitamin D-Related Disorders; Focus on Vitamin D Receptor". Current Drug Metabolism. 19 (12): 1042–1052. doi:10.2174/1389200219666180723143552. PMID 30039758. S2CID 51710351.
- Luderer HF, Demay MB (July 2010). "The vitamin D receptor, the skin and stem cells". J. Steroid Biochem. Mol. Biol. 121 (1–2): 314–6. doi:10.1016/j.jsbmb.2010.01.015. PMID 20138991. S2CID 23876206.
- Lisse TS, Saini V, Zhao H, Luderer HF, Gori F, Demay MB (September 2014). "The Vitamin D Receptor Is Required for Activation of cWnt and Hedgehog Signaling in Keratinocytes". Mol. Endocrinol. 28 (10): 1698–1706. doi:10.1210/me.2014-1043. PMC 4179637. PMID 25180455.
- Guzey M, Takayama S, Reed JC (December 2000). "BAG1L enhances trans-activation function of the vitamin D receptor". J. Biol. Chem. 275 (52): 40749–56. doi:10.1074/jbc.M004977200. PMID 10967105.
- Zhao G, Simpson RU (2010). "Membrane Localization, Caveolin-3 Association and Rapid Actions of Vitamin D Receptor in Cardiac Myocytes". Steroids. 75 (8–9): 555–9. doi:10.1016/j.steroids.2009.12.001. PMC 2885558. PMID 20015453.
- ^ Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG (March 1999). "Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators". Mol. Cell. 3 (3): 361–70. doi:10.1016/S1097-2765(00)80463-3. PMID 10198638.
- ^ Tagami T, Lutz WH, Kumar R, Jameson JL (December 1998). "The interaction of the vitamin D receptor with nuclear receptor corepressors and coactivators". Biochem. Biophys. Res. Commun. 253 (2): 358–63. doi:10.1006/bbrc.1998.9799. PMID 9878542.
- ^ Puccetti E, Obradovic D, Beissert T, Bianchini A, Washburn B, Chiaradonna F, Boehrer S, Hoelzer D, Ottmann OG, Pelicci PG, Nervi C, Ruthardt M (December 2002). "AML-associated translocation products block vitamin D(3)-induced differentiation by sequestering the vitamin D(3) receptor". Cancer Res. 62 (23): 7050–8. PMID 12460926.
- Herdick M, Steinmeyer A, Carlberg C (June 2000). "Antagonistic action of a 25-carboxylic ester analogue of 1alpha, 25-dihydroxyvitamin D3 is mediated by a lack of ligand-induced vitamin D receptor interaction with coactivators". J. Biol. Chem. 275 (22): 16506–12. doi:10.1074/jbc.M910000199. PMID 10748178.
- ^ Zhang C, Baudino TA, Dowd DR, Tokumaru H, Wang W, MacDonald PN (November 2001). "Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription". J. Biol. Chem. 276 (44): 40614–20. doi:10.1074/jbc.M106263200. PMID 11514567.
- He B, Wilson EM (March 2003). "Electrostatic Modulation in Steroid Receptor Recruitment of LXXLL and FXXLF Motifs". Mol. Cell. Biol. 23 (6): 2135–50. doi:10.1128/MCB.23.6.2135-2150.2003. PMC 149467. PMID 12612084.
- ^ Baudino TA, Kraichely DM, Jefcoat SC, Winchester SK, Partridge NC, MacDonald PN (June 1998). "Isolation and characterization of a novel coactivator protein, NCoA-62, involved in vitamin D-mediated transcription". J. Biol. Chem. 273 (26): 16434–41. doi:10.1074/jbc.273.26.16434. PMID 9632709.
- Vidal M, Ramana CV, Dusso AS (April 2002). "Stat1-Vitamin D Receptor Interactions Antagonize 1,25-Dihydroxyvitamin D Transcriptional Activity and Enhance Stat1-Mediated Transcription". Mol. Cell. Biol. 22 (8): 2777–87. doi:10.1128/MCB.22.8.2777-2787.2002. PMC 133712. PMID 11909970.
- Ward JO, McConnell MJ, Carlile GW, Pandolfi PP, Licht JD, Freedman LP (December 2001). "The acute promyelocytic leukemia-associated protein, promyelocytic leukemia zinc finger, regulates 1,25-dihydroxyvitamin D(3)-induced monocytic differentiation of U937 cells through a physical interaction with vitamin D(3) receptor". Blood. 98 (12): 3290–300. doi:10.1182/blood.V98.12.3290. PMID 11719366.
Further reading
- Hosoi T (2002). "". Nippon Rinsho. 60 (Suppl 3): 106–10. PMID 11979895.
- Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP (2004). "Genetics and biology of vitamin D receptor polymorphisms". Gene. 338 (2): 143–56. doi:10.1016/j.gene.2004.05.014. hdl:1765/73442. PMID 15315818. S2CID 11023870.
- Norman AW (2007). "Minireview: vitamin D receptor: new assignments for an already busy receptor". Endocrinology. 147 (12): 5542–8. doi:10.1210/en.2006-0946. PMID 16946007.
- Bollag WB (2007). "Differentiation of human keratinocytes requires the vitamin d receptor and its coactivators". J. Invest. Dermatol. 127 (4): 748–50. doi:10.1038/sj.jid.5700692. PMID 17363957.
- Bugge TH, Pohl J, Lonnoy O, Stunnenberg HG (1992). "RXR alpha, a promiscuous partner of retinoic acid and thyroid hormone receptors". EMBO J. 11 (4): 1409–18. doi:10.1002/j.1460-2075.1992.tb05186.x. PMC 556590. PMID 1314167.
- Goto H, Chen KS, Prahl JM, DeLuca HF (1992). "A single receptor identical with that from intestine/T47D cells mediates the action of 1,25-dihydroxyvitamin D-3 in HL-60 cells". Biochim. Biophys. Acta. 1132 (1): 103–8. doi:10.1016/0167-4781(92)90063-6. PMID 1324736.
- Saijo T, Ito M, Takeda E, Huq AH, Naito E, Yokota I, Sone T, Pike JW, Kuroda Y (1991). "A unique mutation in the vitamin D receptor gene in three Japanese patients with vitamin D-dependent rickets type II: utility of single-strand conformation polymorphism analysis for heterozygous carrier detection". Am. J. Hum. Genet. 49 (3): 668–73. PMC 1683124. PMID 1652893.
- Szpirer J, Szpirer C, Riviere M, Levan G, Marynen P, Cassiman JJ, Wiese R, DeLuca HF (1992). "The Sp1 transcription factor gene (SP1) and the 1,25-dihydroxyvitamin D3 receptor gene (VDR) are colocalized on human chromosome arm 12q and rat chromosome 7". Genomics. 11 (1): 168–73. doi:10.1016/0888-7543(91)90114-T. PMID 1662663.
- Yu XP, Mocharla H, Hustmyer FG, Manolagas SC (1991). "Vitamin D receptor expression in human lymphocytes. Signal requirements and characterization by western blots and DNA sequencing". J. Biol. Chem. 266 (12): 7588–95. doi:10.1016/S0021-9258(20)89488-5. PMID 1850412.
- Malloy PJ, Hochberg Z, Tiosano D, Pike JW, Hughes MR, Feldman D (1991). "The molecular basis of hereditary 1,25-dihydroxyvitamin D3 resistant rickets in seven related families". J. Clin. Invest. 86 (6): 2071–9. doi:10.1172/JCI114944. PMC 329846. PMID 2174914.
- Sone T, Marx SJ, Liberman UA, Pike JW (1991). "A unique point mutation in the human vitamin D receptor chromosomal gene confers hereditary resistance to 1,25-dihydroxyvitamin D3". Mol. Endocrinol. 4 (4): 623–31. doi:10.1210/mend-4-4-623. PMID 2177843.
- Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, Pike JW, Shine J, O'Malley BW (1988). "Cloning and expression of full-length cDNA encoding human vitamin D receptor". Proc. Natl. Acad. Sci. U.S.A. 85 (10): 3294–8. Bibcode:1988PNAS...85.3294B. doi:10.1073/pnas.85.10.3294. PMC 280195. PMID 2835767.
- Hughes MR, Malloy PJ, Kieback DG, Kesterson RA, Pike JW, Feldman D, O'Malley BW (1989). "Point mutations in the human vitamin D receptor gene associated with hypocalcemic rickets". Science. 242 (4886): 1702–5. doi:10.1126/science.2849209. PMID 2849209.
- Rut AR, Hewison M, Kristjansson K, Luisi B, Hughes MR, O'Riordan JL (1995). "Two mutations causing vitamin D resistant rickets: modelling on the basis of steroid hormone receptor DNA-binding domain crystal structures". Clin. Endocrinol. 41 (5): 581–90. doi:10.1111/j.1365-2265.1994.tb01822.x. PMID 7828346. S2CID 40851942.
- Malloy PJ, Weisman Y, Feldman D (1994). "Hereditary 1 alpha,25-dihydroxyvitamin D-resistant rickets resulting from a mutation in the vitamin D receptor deoxyribonucleic acid-binding domain". J. Clin. Endocrinol. Metab. 78 (2): 313–6. doi:10.1210/jcem.78.2.8106618. PMID 8106618.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Yagi H, Ozono K, Miyake H, Nagashima K, Kuroume T, Pike JW (1993). "A new point mutation in the deoxyribonucleic acid-binding domain of the vitamin D receptor in a kindred with hereditary 1,25-dihydroxyvitamin D-resistant rickets". J. Clin. Endocrinol. Metab. 76 (2): 509–12. doi:10.1210/jcem.76.2.8381803. PMID 8381803.
- Kristjansson K, Rut AR, Hewison M, O'Riordan JL, Hughes MR (1993). "Two mutations in the hormone binding domain of the vitamin D receptor cause tissue resistance to 1,25 dihydroxyvitamin D3". J. Clin. Invest. 92 (1): 12–6. doi:10.1172/JCI116539. PMC 293517. PMID 8392085.
- Jurutka PW, Hsieh JC, Nakajima S, Haussler CA, Whitfield GK, Haussler MR (1996). "Human vitamin D receptor phosphorylation by casein kinase II at Ser-208 potentiates transcriptional activation". Proc. Natl. Acad. Sci. U.S.A. 93 (8): 3519–24. Bibcode:1996PNAS...93.3519J. doi:10.1073/pnas.93.8.3519. PMC 39642. PMID 8622969.
- Lin NU, Malloy PJ, Sakati N, al-Ashwal A, Feldman D (1996). "A novel mutation in the deoxyribonucleic acid-binding domain of the vitamin D receptor causes hereditary 1,25-dihydroxyvitamin D-resistant rickets". J. Clin. Endocrinol. Metab. 81 (7): 2564–9. doi:10.1210/jcem.81.7.8675579. PMID 8675579. S2CID 46366654.
External links
- Calcitriol+Receptors at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Nuclear Receptor Resource
- Vitamin D Receptor: Molecule of the Month Archived 2015-10-16 at the Wayback Machine
- IUPHAR: Vitamin D receptor
- Overview of all the structural information available in the PDB for UniProt: P11473 (Vitamin D3 receptor) at the PDBe-KB.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
| PDB gallery | |
|---|---|
|
| Transcription factors and intracellular receptors | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| see also transcription factor/coregulator deficiencies | |||||||||||||||||||||||||||||||
| Vitamin D receptor modulators | |
|---|---|
| VDRTooltip Vitamin D receptor |
|
| |