| Zinc dependent phospholipase C | |||||||||
|---|---|---|---|---|---|---|---|---|---|
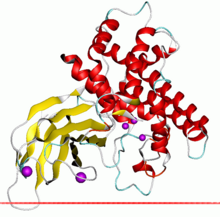 Alpha toxin of Clostridium showing the zinc dependent phospholipase domain in red and the PLAT domain in yellow Alpha toxin of Clostridium showing the zinc dependent phospholipase domain in red and the PLAT domain in yellow | |||||||||
| Identifiers | |||||||||
| Symbol | Zn_dep_PLPC | ||||||||
| Pfam | PF00882 | ||||||||
| InterPro | IPR001531 | ||||||||
| PROSITE | PDOC00357 | ||||||||
| SCOP2 | 1ah7 / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 81 | ||||||||
| OPM protein | 1olp | ||||||||
| CDD | cd11009 | ||||||||
| |||||||||
In molecular biology, zinc-dependent phospholipases C is a family of bacterial phospholipases C enzymes, some of which are also known as alpha toxins.
Bacillus cereus contains a monomeric phospholipase C EC 3.1.4.3 (PLC) of 245 amino-acid residues. Although PLC prefers to act on phosphatidylcholine, it also shows weak catalytic activity with sphingomyelin and phosphatidylinositol. Sequence studies have shown the protein to be similar both to alpha toxin from Clostridium perfringens and Clostridium bifermentans, a phospholipase C involved in haemolysis and cell rupture, and to lecithinase from Listeria monocytogenes, which aids cell-to-cell spread by breaking down the 2-membrane vacuoles that surround the bacterium during transfer.
Each of these proteins is a zinc-dependent enzyme, binding 3 zinc ions per molecule. The enzymes catalyse the conversion of phosphatidylcholine and water to 1,2-diacylglycerol and choline phosphate.
In Bacillus cereus, there are nine residues known to be involved in binding the zinc ions: 5 His, 2 Asp, 1 Glu and 1 Trp. These residues are all conserved in the Clostridium alpha-toxin.
Some examples of this enzyme contain a C-terminal sequence extension that contains a PLAT domain which is thought to be involved in membrane localisation.
References
- ^ Nakamura S, Yamada A, Tsukagoshi N, Udaka S, Sasaki T, Makino S, Little C, Tomita M, Ikezawa H (1988). "Nucleotide sequence and expression in Escherichia coli of the gene coding for sphingomyelinase of Bacillus cereus". Eur. J. Biochem. 175 (2): 213–220. doi:10.1111/j.1432-1033.1988.tb14186.x. PMID 2841128.
- ^ Titball RW, Rubidge T, Hunter SE, Martin KL, Morris BC, Shuttleworth AD, Anderson DW, Kelly DC (1989). "Molecular cloning and nucleotide sequence of the alpha-toxin (phospholipase C) of Clostridium perfringens". Infect. Immun. 57 (2): 367–376. doi:10.1128/IAI.57.2.367-376.1989. PMC 313106. PMID 2536355.
- Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P, Vazquez-Boland JA (1992). "Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread". Infect. Immun. 60 (1): 219–230. doi:10.1128/IAI.60.1.219-230.1992. PMC 257526. PMID 1309513.
- ^ Titball RW, Rubidge T (1990). "The role of histidine residues in the alpha toxin of Clostridium perfringens". FEMS Microbiol. Lett. 56 (3): 261–265. doi:10.1111/j.1574-6968.1988.tb03188.x. PMID 2111259.
- Bateman A, Sandford R (1999). "The PLAT domain: a new piece in the PKD1 puzzle". Curr. Biol. 9 (16): R588–90. doi:10.1016/S0960-9822(99)80380-7. PMID 10469604. S2CID 15018010.
- Ponting CP, Hofmann K, Bork P (August 1999). "A latrophilin/CL-1-like GPS domain in polycystin-1". Curr. Biol. 9 (16): R585–8. doi:10.1016/S0960-9822(99)80379-0. PMID 10469603. S2CID 17252179.
| Enzymes | |
|---|---|
| Activity | |
| Regulation | |
| Classification | |
| Kinetics | |
| Types |
|
This membrane protein–related article is a stub. You can help Misplaced Pages by expanding it. |