This is an old revision of this page, as edited by Evolution and evolvability (talk | contribs) at 02:54, 10 June 2018 (→Biosynthesis: img left). The present address (URL) is a permanent link to this revision, which may differ significantly from the current revision.
Revision as of 02:54, 10 June 2018 by Evolution and evolvability (talk | contribs) (→Biosynthesis: img left)(diff) ← Previous revision | Latest revision (diff) | Newer revision → (diff)

| |||
| |||
| Names | |||
|---|---|---|---|
| IUPAC name Lysine | |||
| Other names 2,6-Diaminohexanoic acid; 2,6-Diammoniohexanoic acid | |||
| Identifiers | |||
| CAS Number | |||
| 3D model (JSmol) | |||
| ChEBI | |||
| ChEMBL | |||
| ChemSpider | |||
| ECHA InfoCard | 100.000.673 | ||
| IUPHAR/BPS | |||
| KEGG | |||
| PubChem CID | |||
| UNII | |||
| CompTox Dashboard (EPA) | |||
InChI
| |||
SMILES
| |||
| Properties | |||
| Chemical formula | C6H14N2O2 | ||
| Molar mass | 146.190 g·mol | ||
| Solubility in water | 1.5 kg/L @ 25 °C | ||
| Pharmacology | |||
| ATC code | B05XB03 (WHO) | ||
| Supplementary data page | |||
| Lysine (data page) | |||
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C , 100 kPa).
| |||
Lysine (symbol Lys or K), encoded by the codons AAA and AAG, is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH3 form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO form under biological conditions), and a side chain lysyl ((CH2)4NH2), classifying it as a charged (at physiological pH), aliphatic amino acid. It is essential in humans, meaning the body cannot synthesize it and thus it must be obtained from the diet.
Lysine is a base, as are arginine and histidine. The ε-amino group often participates in hydrogen bonding and as a general base in catalysis. The ε-ammonium group (NH3) is attached to the fourth carbon from the α-carbon, which is attached to the carboxyl (C=OOH) group.
Common posttranslational modifications include methylation of the ε-amino group, giving methyl-, dimethyl-, and trimethyllysine (the latter occurring in calmodulin); also acetylation, sumoylation, ubiquitination, and hydroxylation – producing the hydroxylysine in collagen and other proteins. O-Glycosylation of hydroxylysine residues in the endoplasmic reticulum or Golgi apparatus is used to mark certain proteins for secretion from the cell. In opsins like rhodopsin and the visual opsins (encoded by the genes OPN1SW, OPN1MW, and OPN1LW), retinaldehyde forms a Schiff base with a conserved lysine residue, and interaction of light with the retinylidene group causes signal transduction in color vision (See visual cycle for details). Deficiencies may cause blindness, as well as many other problems due to its ubiquitous presence in proteins.
Lysine was first isolated by the German biological chemist Ferdinand Heinrich Edmund Drechsel (1843–1897) in 1889 from the protein casein in milk. He named it "lysin". In 1902, the German chemists Emil Fischer and Fritz Weigert determined lysine's chemical structure by synthesizing it.
Biosynthesis
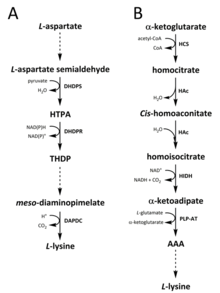
Two different pathways have been identified in nature for the synthesis of lysine. The diaminopimelate (DAP) pathway (Fig. 2A) belongs to the aspartate derived biosynthetic family, which is also involved in the synthesis of threonine, methionine and isoleucine. Whereas the α-aminoadipate (AAA) pathway (Fig. 2B) is part of the glutamate biosynthetic family.
The DAP pathway (Fig. 2A) is found in both prokaryotes and plants and begins with the dihydrodipicolinate synthase (DHDPS) (E.C 4.2.1.52) catalysed condensation reaction between the aspartate derived, L-aspartate semialdehyde, and pyruvate to form (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinic acid (HTPA) (Fig. 2A). The product is then reduced by dihydrodipicolinate reductase (DHDPR) (E.C 1.3.1.26), with NAD(P)H as a proton donor, to yield 2,3,4,5-tetrahydrodipicolinate (THDP) (Fig. 2A). From this point on, there are four pathway variations found in different species, namely the acetylase, aminotransferase, dehydrogenase, and succinylase pathways. Both the acetylase and succinylase variant pathways use four enzyme catalysed steps, the aminotransferase pathway uses two enzymes, and the dehydrogenase pathway uses a single enzyme. These four variant pathways converge at the formation of the penultimate product, meso‑diaminopimelate, which is subsequently enzymatically decarboxylated in an irreversible reaction catalysed by diaminopimelate decarboxylase (DAPDC) (E.C 4.1.1.20) to produce L-lysine (Fig. 2A). The DAP pathway is regulated at multiple levels, including upstream at the enzymes involved in aspartate processing as well as at the initial DHDPS catalysed condensation step. Lysine imparts a strong negative feedback loop on these enzymes and, subsequently, regulates the entire pathway.
The AAA pathway (Fig. 2B) involves the condensation of α-ketoglutarate and acetyl-CoA via the intermediate AAA for the synthesis of L-lysine. This pathway has been shown to be present in several yeast species, as well as protists and higher fungi. It has also been reported that an alternative variant of the AAA route has been found in Thermus thermophilus and Pyrococcus horikoshii, which could indicate that this pathway is more widely spread in prokaryotes than originally proposed. The first and rate-limiting step in the AAA pathway is the condensation reaction between acetyl-CoA and α‑ketoglutarate catalysed by homocitrate-synthase (HCS) (E.C 2.3.3.14) to give the intermediate homocitryl‑CoA, which is hydrolysed by the same enzyme to produce homocitrate (Fig. 2B). Homocitrate is enzymatically dehydrated by homoaconitase (HAc) (E.C 4.2.1.36) to yield cis-homoaconitate. HAc then catalyses a second reaction in which cis-homoaconitate undergoes rehydration to produce homoisocitrate (Fig. 2B). The resulting product undergoes an oxidative decarboxylation by homoisocitrate dehydrogenase (HIDH) (E.C 1.1.1.87) to yield α‑ketoadipate. AAA is then formed via a pyridoxal 5′-phosphate (PLP)-dependent aminotransferase (PLP-AT) (E.C 2.6.1.39), using glutamate as the amino donor (Fig. 2B). From this point on, the AAA pathway differs depending on the kingdom. In fungi, AAA is reduced to α‑aminoadipate-semialdehyde via AAA reductase (E.C 1.2.1.95) in a unique process involving both adenylation and reduction that is activated by a phosphopantetheinyl transferase (E.C 2.7.8.7). Once the semialdehyde is formed, saccharopine reductase (E.C 1.5.1.10) catalyses a condensation reaction with glutamate and NAD(P)H, as a proton donor, and the imine is reduced to produce the penultimate product, saccharopine. The final step of the pathway in fungi involves the saccharopine dehydrogenase (SDH) (E.C 1.5.1.8) catalysed oxidative deamination of saccharopine, resulting in L-lysine. In a variant AAA pathway found in some prokaryotes, AAA is first converted to N‑acetyl-α-aminoadipate, which is phosphorylated and then reductively dephosphorylated to the ε-aldehyde. The aldehyde is then transaminated to N‑acetyl-lysine, which is deacetylated to give L-lysine. However, the enzymes involved in this variant pathway need further validation.
Metabolism
Lysine is metabolised in mammals to give acetyl-CoA, via an initial transamination with α-ketoglutarate. The bacterial degradation of lysine yields cadaverine by decarboxylation.
Allysine is a derivative of lysine, used in the production of elastin and collagen. It is produced by the actions of the enzyme lysyl oxidase on lysine in the extracellular matrix and is essential in the crosslink formation that stabilizes collagen and elastin.
Requirements
The Food and Nutrition Board (FNB) of the U.S. Institute of Medicine set Recommended Dietary Allowances (RDAs) for essential amino acids in 2002. For lysine, for adults 19 years and older, 38 mg/kg body weight/day.
Synthesis
Synthetic, racemic lysine has long been known. A practical synthesis starts from caprolactam. Industrially, L-lysine is usually manufactured by a fermentation process using Corynebacterium glutamicum; production exceeds 600,000 tons a year.
L-lysine HCl is used as a dietary supplement, providing 80.03% L-lysine. As such, 1 g of L-lysine is contained in 1.25 g of L-lysine HCl.
Dietary sources
The nutritional requirement per day, in milligrams of lysine per kilogram of body weight, is: infants (3–4 months) 103 mg/kg, children (2 years) 64 mg/kg, older children (10–12 years) 44 to 60 mg/kg, adults 12 mg/kg. For a 70 kg adult, 12 milligrams of lysine per kilogram of body weight is 0.84 grams of lysine. Recommendations for adults have been revised upwards to 30 mg/kg.
Good sources of lysine are high-protein foods such as eggs, meat (specifically red meat, lamb, pork, and poultry), soy, beans and peas, cheese (particularly Parmesan), and certain fish (such as cod and sardines).
Lysine is the limiting amino acid (the essential amino acid found in the smallest quantity in the particular foodstuff) in most cereal grains, but is plentiful in most pulses (legumes). A vegetarian or low animal protein diet can be adequate for protein, including lysine, if it includes both cereal grains and legumes, but there is no need for the two food groups to be consumed in the same meals.
A food is considered to have sufficient lysine if it has at least 51 mg of lysine per gram of protein (so that the protein is 5.1% lysine). Foods containing significant proportions of lysine include:
| Food | Lysine (% of protein) |
|---|---|
| Fish | 9.19% |
| Beef, ground, 90% lean/10% fat, cooked | 8.31% |
| Chicken, roasting, meat and skin, cooked, roasted | 8.11% |
| Azuki bean (adzuki beans), mature seeds, raw | 7.53% |
| Milk, non-fat | 7.48% |
| Soybean, mature seeds, raw | 7.42% |
| Egg, whole, raw | 7.27% |
| Pea, split, mature seeds, raw | 7.22% |
| Lentil, pink, raw | 6.97% |
| Kidney bean, mature seeds, raw | 6.87% |
| Chickpea, (garbanzo beans, Bengal gram), mature seeds, raw | 6.69% |
| Navy bean, mature seeds, raw | 5.73% |
Properties
L-Lysine plays a major role in calcium absorption; building muscle protein; recovering from surgery or sports injuries; and the body's production of hormones, enzymes, and antibodies.
Modifications
Lysine can be modified through acetylation (acetyllysine), methylation (methyllysine), ubiquitination, sumoylation, neddylation, biotinylation, pupylation, and carboxylation, which tends to modify the function of the protein of which the modified lysine residue(s) are a part.
Clinical significance
A review cited studies showing that lysine supplementation can decrease herpes simplex cold sore outbreaks and reduce healing time. Original article published at 1978.
However, at 1984 and later the controlled researches don't confirm this for humans and animals.
An authoritative Cochrane Review published in 2015 concluded there is insufficient evidence that lysine supplementation is effective against herpes simplex virus; it has not been approved by the FDA for herpes simplex suppression.
Use of lysine in animal feed
Lysine production for animal feed is a major global industry, reaching in 2009 almost 700,000 tonnes for a market value of over €1.22 billion. Lysine is an important additive to animal feed because it is a limiting amino acid when optimizing the growth of certain animals such as pigs and chickens for the production of meat. Lysine supplementation allows for the use of lower-cost plant protein (maize, for instance, rather than soy) while maintaining high growth rates, and limiting the pollution from nitrogen excretion. In turn, however, phosphate pollution is a major environmental cost when corn is used as feed for poultry and swine.
Lysine is industrially produced by microbial fermentation, from a base mainly of sugar. Genetic engineering research is actively pursuing bacterial strains to improve the efficiency of production and allow lysine to be made from other substrates.
In popular culture
The 1993 film Jurassic Park (based on the 1990 Michael Crichton novel of the same name) features dinosaurs that were genetically altered so that they could not produce lysine. This was known as the "lysine contingency" and was supposed to prevent the cloned dinosaurs from surviving outside the park, forcing them to be dependent on lysine supplements provided by the park's veterinary staff. In reality, no animals are capable of producing lysine (it is an essential amino acid).
Lysine is the favorite amino acid of the character Sheldon Cooper in the television show, The Big Bang Theory. It was mentioned in season 2, episode 13, "The Friendship Algorithm".
In 1996, lysine became the focus of a price-fixing case, the largest in United States history. The Archer Daniels Midland Company paid a fine of US$100 million, and three of its executives were convicted and served prison time. Also found guilty in the price-fixing case were two Japanese firms (Ajinomoto, Kyowa Hakko) and a South Korean firm (Sewon). Secret video recordings of the conspirators fixing lysine's price can be found online or by requesting the video from the U.S. Department of Justice, Antitrust Division. This case served as the basis of the movie The Informant!, and a book of the same title.
The 2002 album Mastered by Guy at The Exchange by Max Tundra features a song called "Lysine".
See also
References
- IUPAC-IUBMB Joint Commission on Biochemical Nomenclature. "Nomenclature and Symbolism for Amino Acids and Peptides". Recommendations on Organic & Biochemical Nomenclature, Symbols & Terminology etc. Retrieved 17 May 2007.
- Lysine. The Biology Project, Department of Biochemistry and Molecular Biophysics, University of Arizona.
- Drechsel, E. (1889). "Zur Kenntniss der Spaltungsprodukte des Caseïns" [ to knowledge of the cleavage products of casein]. Journal für Praktische Chemie. 2nd series (in German). 39: 425–429. On p. 428, Drechsel presented an empirical formula for the chloroplatinate salt of lysine – C8H16N2O2Cl2•PtCl4 + H2O – but he later admitted that this formula was wrong because the salt's crystals contained ethanol instead of water. See: Drechsel, E. (1891) "Der Abbau der Eiweissstoffe" , Archiv für Anatomie und Physiologie, 248–278. §2. Drechsel, E. "Zur Kenntniss der Spaltungsproducte des Caseïns" ( to knowledge of the cleavage products of casein), pp. 254–260. From p. 256: " … die darin enthaltene Base hat die Formel C6H14N2O2. Der anfängliche Irrthum ist dadurch veranlasst worden, dass das Chloroplatinat nicht, wie angenommen ward, Krystallwasser, sondern Krystallalkohol enthält, … " ( … the base contained therein has the formula C6H14N2O2. The initial error was caused by the chloroplatinate containing not water in the crystal (as was assumed), but ethanol … )
- Drechsel, E. (1891) "Der Abbau der Eiweissstoffe" , Archiv für Anatomie und Physiologie, 248–278. §4. Fischer, Ernst (1891) "Ueber neue Spaltungsproducte des Leimes" (On new cleavage products of gelatin), pp. 465–469. From p. 469: " … die Base C6H14N2O2, welche mit dem Namen Lysin bezeichnet werden mag, … " ( … the base C6H14N2O2, which may be designated with the name "lysine", … )
- Fischer, Emil; Weigert, Fritz (1902). "Synthese der α,ε – Diaminocapronsäure (Inactives Lysin)" [Synthesis of α,ε-diaminohexanoic acid ( inactive lysine)]. Berichte der Deutschen Chemischen Gesellschaft (in German). 35: 3772–3778.
- ^ Hudson, Andre' O.; Bless, Christine; Macedo, Polliana; Chatterjee, Siba P.; Singh, Bijay K.; Gilvarg, Charles; Leustek, Thomas. "Biosynthesis of lysine in plants: evidence for a variant of the known bacterial pathways". Biochimica et Biophysica Acta (BBA) - General Subjects. 1721 (1–3): 27–36. doi:10.1016/j.bbagen.2004.09.008.
- Velasco, A. M.; Leguina, J. I.; Lazcano, A. (1 October 2002). "Molecular Evolution of the Lysine Biosynthetic Pathways". Journal of Molecular Evolution. 55 (4): 445–449. doi:10.1007/s00239-002-2340-2. ISSN 0022-2844.
- Miyazaki, Takashi; Miyazaki, Junichi; Yamane, Hisakazu; Nishiyama, Makoto (2004). "α-Aminoadipate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus". Microbiology. 150 (7): 2327–2334. doi:10.1099/mic.0.27037-0.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Xu, Hengyu; Andi, Babak; Qian, Jinghua; West, Ann H.; Cook, Paul F. (1 September 2006). "The α-aminoadipate pathway for lysine biosynthesis in fungi". Cell Biochemistry and Biophysics. 46 (1): 43–64. doi:10.1385/cbb:46:1:43. ISSN 1085-9195.
- Atkinson, Sarah C.; Dogovski, Con; Downton, Matthew T.; Czabotar, Peter E.; Dobson, Renwick C. J.; Gerrard, Juliet A.; Wagner, John; Perugini, Matthew A. (1 March 2013). "Structural, kinetic and computational investigation of Vitis vinifera DHDPS reveals new insight into the mechanism of lysine-mediated allosteric inhibition". Plant Molecular Biology. 81 (4–5): 431–446. doi:10.1007/s11103-013-0014-7. ISSN 0167-4412.
- Griffin, Michael D. W.; Billakanti, Jagan M.; Wason, Akshita; Keller, Sabrina; Mertens, Haydyn D. T.; Atkinson, Sarah C.; Dobson, Renwick C. J.; Perugini, Matthew A.; Gerrard, Juliet A. (5 July 2012). "Characterisation of the First Enzymes Committed to Lysine Biosynthesis in Arabidopsis thaliana". PLOS ONE. 7 (7): e40318. doi:10.1371/journal.pone.0040318. ISSN 1932-6203.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Soares da Costa, Tatiana P.; Muscroft-Taylor, Andrew C.; Dobson, Renwick C.J.; Devenish, Sean R.A.; Jameson, Geoffrey B.; Gerrard, Juliet A. "How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?". Biochimie. 92 (7): 837–845. doi:10.1016/j.biochi.2010.03.004.
- Soares da Costa, Tatiana P.; Christensen, Janni B.; Desbois, Sebastien; Gordon, Shane E.; Gupta, Ruchi; Hogan, Campbell J.; Nelson, Tao G.; Downton, Matthew T.; Gardhi, Chamodi K. Quaternary Structure Analyses of an Essential Oligomeric Enzyme. pp. 205–223. doi:10.1016/bs.mie.2015.06.020.
- Muscroft-Taylor, Andrew C.; Costa, Tatiana P. Soares da; Gerrard, Juliet A. "New insights into the mechanism of dihydrodipicolinate synthase using isothermal titration calorimetry". Biochimie. 92 (3): 254–262. doi:10.1016/j.biochi.2009.12.004.
- Christensen, Janni B.; Costa, T. P. Soares da; Faou, Pierre; Pearce, F. Grant; Panjikar, Santosh; Perugini, Matthew A. (15 November 2016). "Structure and Function of Cyanobacterial DHDPS and DHDPR". Scientific Reports. 6 (1). doi:10.1038/srep37111. ISSN 2045-2322.
- McCoy, Andrea J.; Adams, Nancy E.; Hudson, André O.; Gilvarg, Charles; Leustek, Thomas; Maurelli, Anthony T. (21 November 2006). "l,l-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine". Proceedings of the National Academy of Sciences. 103 (47): 17909–17914. doi:10.1073/pnas.0608643103.
- Hudson, André O.; Gilvarg, Charles; Leustek, Thomas (1 May 2008). "Biochemical and Phylogenetic Characterization of a Novel Diaminopimelate Biosynthesis Pathway in Prokaryotes Identifies a Diverged Form of ll-Diaminopimelate Aminotransferase". Journal of Bacteriology. 190 (9): 3256–3263. doi:10.1128/jb.01381-07. ISSN 0021-9193. PMID 18310350.
- Peverelli, Martin G.; Perugini, Matthew A. "An optimized coupled assay for quantifying diaminopimelate decarboxylase activity". Biochimie. 115: 78–85. doi:10.1016/j.biochi.2015.05.004.
- ^ Soares da Costa, Tatiana P.; Costa, Tatiana P. Soares da; Desbois, Sebastien; Dogovski, Con; Gorman, Michael A.; Ketaren, Natalia E.; Paxman, Jason J.; Siddiqui, Tanzeela; Zammit, Leanne M. "Structural Determinants Defining the Allosteric Inhibition of an Essential Antibiotic Target". Structure. 24 (8): 1282–1291. doi:10.1016/j.str.2016.05.019.
{{cite journal}}: no-break space character in|last=at position 10 (help) - ^ Jander, Georg; Joshi, Vijay (1 January 2009). "Aspartate-Derived Amino Acid Biosynthesis in Arabidopsis thaliana". The Arabidopsis Book: e0121. doi:10.1199/tab.0121.
- Andi, Babak; West, Ann H.; Cook, Paul F. (1 September 2004). "Kinetic Mechanism of Histidine-Tagged Homocitrate Synthase from Saccharomyces cerevisiae". Biochemistry. 43 (37): 11790–11795. doi:10.1021/bi048766p. ISSN 0006-2960.
- Bhattacharjee, J. K. (1985). "alpha-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes". Critical Reviews in Microbiology. 12 (2): 131–151. doi:10.3109/10408418509104427. ISSN 1040-841X. PMID 3928261.
- Bhattacharjee, J. K.; Strassman, M. (25 May 1967). "Accumulation of tricarboxylic acids related to lysine biosynthesis in a yeast mutant". The Journal of Biological Chemistry. 242 (10): 2542–2546. ISSN 0021-9258. PMID 6026248.
- GAILLARDIN, Claude M.; RIBET, Anne-Marie; HESLOT, Henri. "Wild-Type and Mutant Forms of Homoisocitric Dehydrogenase in the Yeast Saccharomycopsis lipolytica". European Journal of Biochemistry. 128 (2–3): 489–494. doi:10.1111/j.1432-1033.1982.tb06991.x.
- Jaklitsch, W. M.; Kubicek, C. P. (1 July 1990). "Homocitrate synthase from Penicillium chrysogenum. Localization, purification of the cytosolic isoenzyme, and sensitivity to lysine". The Biochemical Journal. 269 (1): 247–253. ISSN 0264-6021. PMC 1131560. PMID 2115771.
{{cite journal}}: CS1 maint: PMC format (link) - Ye, Z. H.; Bhattacharjee, J. K. (1 December 1988). "Lysine biosynthesis pathway and biochemical blocks of lysine auxotrophs of Schizosaccharomyces pombe". Journal of Bacteriology. 170 (12): 5968–5970. doi:10.1128/jb.170.12.5968-5970.1988. ISSN 0021-9193. PMID 3142867.
- Kobashi, N.; Nishiyama, M.; Tanokura, M. (March 1999). "Aspartate kinase-independent lysine synthesis in an extremely thermophilic bacterium, Thermus thermophilus: lysine is synthesized via alpha-aminoadipic acid not via diaminopimelic acid". Journal of Bacteriology. 181 (6): 1713–1718. ISSN 0021-9193. PMID 10074061.
- Kosuge, T.; Hoshino, T. (1999). "The alpha-aminoadipate pathway for lysine biosynthesis is widely distributed among Thermus strains". Journal of Bioscience and Bioengineering. 88 (6): 672–675. ISSN 1389-1723. PMID 16232683.
- ^ Nishida, H. "A Prokaryotic Gene Cluster Involved in Synthesis of Lysine through the Amino Adipate Pathway: A Key to the Evolution of Amino Acid Biosynthesis". Genome Research. 9 (12): 1175–1183. doi:10.1101/gr.9.12.1175.
- ^ Nishida, H.; Nishiyama, M. (September 2000). "What is characteristic of fungal lysine synthesis through the alpha-aminoadipate pathway?". Journal of Molecular Evolution. 51 (3): 299–302. ISSN 0022-2844. PMID 11029074.
- Zabriskie, T. Mark; Jackson, Michael D. (1 January 2000). "Lysine biosynthesis and metabolism in fungi". Natural Product Reports. 17 (1): 85–97. doi:10.1039/a801345d. ISSN 1460-4752.
- Institute of Medicine (2002). "Protein and Amino Acids". Dietary Reference Intakes for Energy, Carbohydrates, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids. Washington, DC: The National Academies Press. pp. 589–768.
- Braun, J. V. (1909). "Synthese des inaktiven Lysins aus Piperidin". Berichte der deutschen chemischen Gesellschaft. 42: 839–846. doi:10.1002/cber.190904201134.
- Eck, J. C.; Marvel, C. S. (1943). "dl-Lysine Hydrochlorides" (PDF). Organic Syntheses, Collected. 2: 374. Archived from the original (PDF) on 27 September 2007.
{{cite journal}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help) - Pfefferle, W.; Möckel, B.; Bathe, B.; Marx, A. (2003). "Biotechnological manufacture of lysine". Advances in Biochemical Engineering/Biotechnology. Advances in Biochemical Engineering/Biotechnology. 79: 59–112. doi:10.1007/3-540-45989-8_3. ISBN 978-3-540-43383-5. PMID 12523389.
- "Dietary Supplement Database: Blend Information (DSBI)".
L-LYSINE HCL 10000820 80.03% lysine
- United Nations Food; Agriculture Organization : Agriculture and Consumer Protection. "Energy and protein requirements: 5.6 Requirements for essential amino acids". Retrieved 10 October 2010.
{{cite web}}: Unknown parameter|lastauthoramp=ignored (|name-list-style=suggested) (help) - FAO/WHO/UNU (2007). "PROTEIN AND AMINO ACID REQUIREMENTS IN HUMAN NUTRITION" (PDF). WHO Press., page 150-152
- University of Maryland Medical Center. "Lysine". Retrieved 30 December 2009.
- Young VR, Pellett PL (1994). "Plant proteins in relation to human protein and amino acid nutrition" (PDF). American Journal of Clinical Nutrition. 59 (5 Suppl): 1203S – 1212S. doi:10.1093/ajcn/59.5.1203s. PMID 8172124.
- Institute of Medicine of the National Academies. "Dietary Reference Intakes for Macronutrients". p. 589. Retrieved 29 October 2017.
- Sadoul K, Boyault C, Pabion M, Khochbin S (February 2008). "Regulation of protein turnover by acetyltransferases and deacetylases". Biochimie. 90 (2): 306–12. doi:10.1016/j.biochi.2007.06.009. PMID 17681659.
- Gaby AR (2006). "Natural remedies for Herpes simplex". Altern Med Rev. 11 (2): 93–101. PMID 16813459.
- Griffith RS, Norins AL, Kagan C. (1978). "A multicentered study of lysine therapy in Herpes simplex infection". Dermatologica. 156 (5): 257–267. doi:10.1159/000250926. PMID 640102.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - John J. DiGiovanna, MD; Harvey Blank, MD (1984). "Failure of Lysine in Frequently Recurrent Herpes Simplex Infection: Treatment and Prophylaxis". Dermatology. 120 (1): 48–51. doi:10.1001/archderm.1984.01650370054010. Retrieved 1 May 2018.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Sebastiaan Bol, Evelien M Bunnik (2015). "Lysine supplementation is not effective for the prevention or treatment of feline herpesvirus 1 infection in cats: A systematic review". BMC Veterinary Research. 11 (1): 284. doi:10.1186/s12917-015-0594-3.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Ching-Chi Chi1, Shu-Hui Wang, Finola M Delamere, Fenella Wojnarowska, Mathilde C Peters, Preetha P Kanjirath (2015). "Interventions for prevention of herpes simplex labialis (cold sores on the lips)". Cochrane Database of Systematic Reviews (8): CD010095. doi:10.1002/14651858.CD010095.pub2. PMID 26252373. CD010095.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - Drugs.com. "Herpes Simplex, Suppression Medications". Retrieved 19 May 2014.
- ^ "AllAboutFeed – News: Norwegian granted for improving lysine production process". archive.org. 11 March 2012. Retrieved 14 April 2018.
- Toride Y. "Lysine and other amino acids for feed: production and contribution to protein utilization in animal feeding". Retrieved 25 January 2011.
- Abelson, Philip (March 1999). "A Potential Phosphate Crisis". Science. 283 (5410): 2015. doi:10.1126/science.283.5410.2015. PMID 10206902.
- Coyne, Jerry A. (10 October 1999). "The Truth Is Way Out There". The New York Times. Retrieved 6 April 2008.
- Wu, G (2009). "Amino acids: Metabolism, functions, and nutrition". Amino Acids. 37 (1): 1–17. doi:10.1007/s00726-009-0269-0. PMID 19301095.
- M3ta1 (28 February 2011). "Sheldon's Favourite Amino Acid". Retrieved 14 April 2018 – via YouTube.
{{cite web}}: CS1 maint: numeric names: authors list (link) - Connor, J.M.; "Global Price Fixing" 2nd Ed. Springer-Verlag: Heidelberg, 2008. ISBN 978-3-540-78669-6.
- Eichenwald, Kurt.; "The Informant: a true story" Broadway Books: New York, 2000. ISBN 0-7679-0326-9.
Sources
- Much of the information in this article has been translated from German Misplaced Pages.
- Lide, D. R., ed. (2002). CRC Handbook of Chemistry and Physics (83rd ed.). Boca Raton, Florida: CRC Press. ISBN 0-8493-0483-0.
| Encoded (proteinogenic) amino acids | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| General topics |  | ||||||||||
| By properties |
| ||||||||||

