| Alpha | |
 | |
| General details | |
|---|---|
| WHO Designation | Alpha |
| Lineage | B1.1.1.7 |
| First detected | Kent, England |
| Date reported | November 2020; 4 years ago (2020-11) |
| Status | Variant of concern |
| Symptoms | |
| |
| Cases map | |
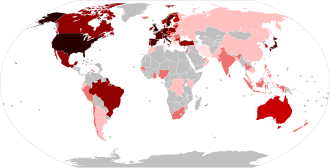 Legend: 10,000+ confirmed sequences 5,000–9,999 confirmed sequences 1,000–4,999 confirmed sequences 500–999 confirmed sequences 100–499 confirmed sequences 2–99 confirmed sequences 1 confirmed sequence None or no data available | |
| Major variants | |
| Omicron (B.1.1.529) | |
The Alpha variant (B.1.1.7) was a SARS-CoV-2 variant of concern. It was estimated to be 40–80% more transmissible than the wild-type SARS-CoV-2 (with most estimates occupying the middle to higher end of this range). Scientists more widely took note of this variant in early December 2020, when a phylogenetic tree showing viral sequences from Kent, United Kingdom looked unusual.
The variant began to spread quickly by mid-December, around the same time as infections surged. This increase is thought to be at least partly because of one or more mutations in the virus' spike protein. The variant was also notable for having more mutations than normally seen. By January 2021, more than half of all genomic sequencing of SARS-CoV-2 was carried out in the UK. This gave rise to questions as to how many other important variants were circulating around the world undetected.
On 2 February 2021, Public Health England reported that they had detected " limited number of B.1.1.7 VOC-202012/01 genomes with E484K mutations", which they dubbed Variant of Concern 202102/02 (VOC-202102/02). One of the mutations (N501Y) was also present in Beta variant and Gamma variant. On 31 May 2021, the World Health Organization announced that the Variant of Concern would be labelled "Alpha" for use in public communications.
The Alpha variant disappeared in late 2021 as a result of competition from even more infectious variants. In March 2022, the World Health Organization changed its designation to "previously circulating variant of concern".
Classification
Names
The variant is known by several names. Outside the UK it is sometimes referred to as the UK variant or British variant or English variant, despite the existence of other, less common, variants first identified in UK, such as Eta variant (lineage B.1.525). Within the UK, it is commonly referred to as the Kent variant after Kent, where the variant was found.
In scientific use, the variant had originally been named the first Variant Under Investigation in December 2020 (VUI – 202012/01) by Public Health England (PHE), but was reclassified to a Variant of Concern (Variant of Concern 202012/01, abbreviated VOC-202012/01) by Meera Chand and her colleagues in a report published by PHE on 21 December 2020. In a report written on behalf of COVID-19 Genomics UK (COG-UK) Consortium, Andrew Rambaut and his co-authors, using the Phylogenetic Assignment of Named Global Outbreak Lineages (pangolin) tool, dubbed it lineage B.1.1.7, while Nextstrain dubbed the variant 20I/501Y.V1.
The name VOC-202102/02 refers to the variant with the E484K mutation (see below).
Genetic profile
-
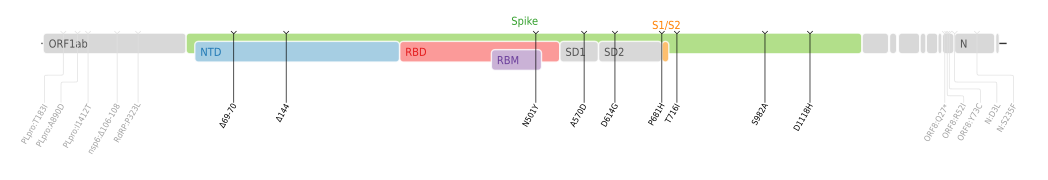 Amino acid mutations of SARS-CoV-2 Alpha variant plotted on a genome map of SARS-CoV-2 with a focus on Spike.
Amino acid mutations of SARS-CoV-2 Alpha variant plotted on a genome map of SARS-CoV-2 with a focus on Spike.
| Gene | Nucleotide | Amino acid |
|---|---|---|
| ORF1ab | C3267T | T1001I |
| C5388A | A1708D | |
| T6954C | I2230T | |
| 11288–11296 deletion | SGF 3675–3677 deletion | |
| Spike | 21765–21770 deletion | HV 69–70 deletion |
| 21991–21993 deletion | Y144 deletion | |
| A23063T | N501Y | |
| C23271A | A570D | |
| C23604A | P681H | |
| C23709T | T716I | |
| T24506G | S982A | |
| G24914C | D1118H | |
| ORF8 | C27972T | Q27stop |
| G28048T | R52I | |
| A28111G | Y73C | |
| N | 28280 GAT→CTA | D3L |
| C28977T | S235F | |
| Source: Chand et al., table 1 (p. 5) | ||
Mutations in SARS-CoV-2 are common: over 4,000 mutations have been detected in its spike protein alone, according to the COVID-19 Genomics UK (COG-UK) Consortium.
VOC-202012/01 is defined by 23 mutations: 14 non-synonymous mutations, 3 deletions, and 6 synonymous mutations (i.e., there are 17 mutations that change proteins and six that do not).
Symptoms and signs
Imperial College London investigated over a million people in England while the Alpha variant was dominant and discovered a wide range of further symptoms linked to Covid. "Chills, loss of appetite, headache and muscle aches" were most common in infected people, as well as classic symptoms.
Diagnosis
Several rapid antigen tests for SARS-CoV-2 are in widespread use globally for COVID-19 diagnostics. They are believed to be useful in stopping the chain of transmission of the virus by providing the means to rapidly identify large numbers of cases as part of a mass-testing program. Following the emergence of VOC-202012/01, there was initially concern that rapid tests might not detect it, but Public Health England determined that rapid tests evaluated and used in the United Kingdom detected the variant.
Prevention
See also: Oxford–AstraZeneca COVID-19 vaccine § Effectiveness, Pfizer–BioNTech COVID-19 vaccine § Effectiveness, Moderna COVID-19 vaccine § Effectiveness, Novavax COVID-19 vaccine § Efficacy, Sputnik V COVID-19 vaccine § Variants, CoronaVac § Variants, Covaxin § Variants, and Abdala (vaccine) § EfficacyBy late 2020, several COVID-19 vaccines were being deployed or under development.
However, as further mutations occurred, concerns were raised as to whether vaccine development would need to be altered. SARS-CoV-2 does not mutate as quickly as, for example, influenza viruses, and the new vaccines that had proved effective by the end of 2020 are types that can be adjusted if necessary. As of the end of 2020, German, British, and American health authorities and experts believe that existing vaccines will be as effective against VOC-202012/01 as against previous variants.
On 18 December, NERVTAG determined "that there are currently insufficient data to draw any conclusion on... ntigenic escape".
As of 20 December 2020, Public Health England confirmed there is "no evidence" to suggest that the new variant would be resistant to the Pfizer–BioNTech vaccine currently being used in the UK's vaccination programme, and that people should still be protected.
E484K mutation
On 2 February 2021, Public Health England reported that they had detected " limited number of B.1.1.7 VOC-202012/01 genomes with E484K mutations", which is also present in the Beta and Gamma variants; a mutation which may reduce vaccine effectiveness. On 9 February 2021, it became known that some 76 cases with the E484K mutation had been detected, principally in Bristol, but with a genomically distinct group in Liverpool also carrying the mutation. A week later a Research and analysis report from PHE gave a total of 77 confirmed and probable cases involving the E484K mutation across the UK, in two variants, VUI-202102/01 and VOC-202102/02, the latter described as 'B.1.1.7 with E484K'.
On 5 March 2021, it was reported that a B.1.1.7 lineage with the E484K mutation has been detected in two US patients (in Oregon and New York states). Researchers thought that the E484K mutation in the Oregon variant arose independently.
Characteristics
Transmissibility
The transmissibility of the Alpha variant (lineage B.1.1.7) had generally been found to be substantially higher than that of pre-existing SARS-CoV-2 variants. The variant was discovered by a team of scientists at COG-UK whose initial results found transmissibility was 70% (50-100%) higher. A study by the Centre for the Mathematical Modelling of Infectious Diseases at the London School of Hygiene & Tropical Medicine reported that the variant was 43 to 90% (range of 95% credible intervals, 38 to 130%) more transmissible than pre-existing variants in the United Kingdom, depending on the method used to assess increases in transmissibility. Similar increases in the transmissibility of lineage B.1.1.7 were measured in Denmark, Switzerland, and the United States. Furthermore, a simple model to account for the rapid rise of lineage B.1.1.7 in several countries and the world found that the variant is 50% more transmissible than the local wild type in these three countries and across the world as whole. Another study concluded that it was 75% (70%–80%) more transmissible in the UK between October and November 2020. A later study suggested that these earlier estimates overestimated the transmissibility of the variant and that the transmissibility increase was on the lower ends of these ranges.
The Dutch Ministry of Health, Welfare and Sport calculated, based on genome sequencing of positive cases, each week the transmissibility rate of the variant compared to the local wildtype, and found it to fluctuate between 28%-47% higher during the first six weeks of 2021. The Danish Statens Serum Institut in comparison calculated it to be 55% (48%–62%) more transmissible in Denmark based upon the observed development of its relative frequency from 4 January to 12 February 2021. The Institute of Social and Preventive Medicine (ISPM) under University of Bern, calculated the transmissibility of lineage B.1.1.7 based on the weekly development of its observed fraction of all Covid-19 positives during the entire pandemic, and found for 95% confidence intervals under the assumption of a wildtype reproduction number Rw≈1 and an exponentially generation time of 5.2 days, that transmissibility was: 52% (45%–60%) higher when compared to the wildtype in Denmark and 51% (42%-60%) higher when compared to the wildtype in Switzerland.
On 18 December 2020—early on in the risk assessment of the variant—the UK scientific advisory body New and Emerging Respiratory Virus Threats Advisory Group (NERVTAG) concluded that they had moderate confidence that VOC-202012/01 was substantially more transmissible than other variants, but that there were insufficient data to reach any conclusion on underlying mechanism of increased transmissibility (e.g. increased viral load, tissue distribution of virus replication, serial interval etc.), the age distribution of cases, or disease severity. Data seen by NERVTAG indicated that the relative reproduction number ("multiplicative advantage") was determined to be 1.74—i.e., the variant is 74% more transmissible—assuming a 6.5-day generational interval. It was demonstrated that the variant grew fast exponentially with respect to the other variants. The variant out-competed the ancestral variant by a factor of every two weeks. Another group came to similar conclusions, generating a replicative advantage, independent of "protective measures", of 2.24 per generation of 6.73 days, out-competing the ancestral variant by every two weeks. Similarly, in Ireland, the variant—as indicated by the missing S gene detection (S-gene target failure ), which historically was rare—went from 16.3% to 46.3% of cases in two weeks. This demonstrates, based on the statistics of 116 positive samples, that the variant had a relative higher growth by a factor of , when compared to the average growth for all other variants by the end of this two week period. The variant became the dominant variant in London, East of England and the South East from low levels in one to two months. A surge of SARS-CoV-2 infections around the start of the new year is seen as being the result of the elevated transmissibility of the variant, while the other variants were in decline.
One of the most important changes in lineage B.1.1.7 seems to be N501Y, a change from asparagine (N) to tyrosine (Y) in amino-acid position 501. This is because of its position inside the spike protein's receptor-binding domain (RBD)—more specifically inside the receptor-binding motif (RBM), a part of the RBD—which binds human ACE2. Mutations in the RBD can change antibody recognition and ACE2 binding specificity and lead to the virus becoming more infectious;. Chand et al. concluded that "t is highly likely that N501Y affects the receptor-binding affinity of the spike protein, and it is possible that this mutation alone or in combination with the deletion at 69/70 in the N-terminal domain (NTD) is enhancing the transmissibility of the virus". In early 2021 a peer-reviewed paper found that the mentioned HV 69–70 deletion in vitro "appeared to have two-fold higher infectivity over a single round of infection compared to [wild-type SARS-CoV-2]" in lentiviral SARS-CoV-2 pseudoviruses.
Using In Silico approach, Shahhosseini et al. demonstrated that the Y501 mutation in lineage B.1.1.7 forms a shorter H-bond (length of 2.94 Å) than its counterpart in the wild type (WT) variant residue N501, indicating that in lineage B.1.1.7 the RBD and hACE2 have a stable interaction. Furthermore, the Y501 mutation in lineage B.1.1.7 contributes more negatively to Binding Free Energy (BFE) (-7.18 kcal/mol) than its counterpart in the WT variant residue N501 (-2.92 kcal/mol). As a result of combining BFE and molecular interaction results, the N501Y mutation in RBD strengthens binding affinity of SARS-CoV-2 RBD to hACE2.
In a detailed affinity and kinetic analysis of the interaction between the Spike RBD and ACE2, the N501Y mutation was found to significantly enhance the binding affinity between the RBD and ACE2 by approximately 10-fold, resulting from a 1.8-fold increase in the association rate constant (kon) and a 7-fold decrease in the dissociation rate constant (koff).
Virulence
Matched cohort studies of the Alpha variant (lineage B.1.1.7) found higher mortality rate than earlier circulating variants overall, but not in hospitalised patients. An ecological study found no difference in mortality overall.
Initially, NERVTAG said on 18 December 2020 that there were insufficient data to reach a conclusion regarding disease severity. At prime minister Boris Johnson's briefing the following day, officials said that there was "no evidence" as of that date that the variant caused higher mortality or was affected differently by vaccines and treatments; Vivek Murthy agreed with this. Susan Hopkins, the joint medical adviser for the NHS Test and Trace and Public Health England (PHE), declared in mid-December 2020: "There is currently no evidence that this strain causes more severe illness, although it is being detected in a wide geography, especially where there are increased cases being detected." Around a month later, however—on 22 January 2021—Johnson said that "there is some evidence that the new variant ... may be associated with a higher degree of mortality", though Sir Patrick Vallance, the government's Chief Scientific Advisor, stressed that there is not yet enough evidence to be fully certain of this.
In a paper analysing twelve different studies on lineage B.1.1.7 death rate relative to other lineages, it was found to have a higher death rate (71% according to LSHTM, 70% according to University of Exeter, 65% according to Public Health England, and 36% according to Imperial College London), and NERVTAG concluded: "Based on these analyses, it is likely that infection with VOC B.1.1.7 is associated with an increased risk of hospitalisation and death compared to infection with non-VOC viruses". Results of the death studies were associated with some high uncertainty and confidence intervals, because of a limited sample size related to the fact that UK only analysed the VOC status for 8% of all COVID-19 deaths.
A UK case-control study for 54,906 participants, testing positive for SARS-CoV-2 between 1 October 2020 and 29 January 2021 in the community setting (not including vulnerable persons from care centres and other public institutions), reported that patients infected with the Alpha variant (VOC 202012/1) had a hazard ratio for death within 28 days of testing of 1.64 (95% confidence interval 1.32-2.04), as compared with matched patients positive for other variants of SARS-CoV-2. Also in the UK, a survival analysis of 1,146,534 participants testing positive for SARS-CoV-2 between 1 November 2020 and 14 February 2021, including individuals in the community and in care and nursing homes, found a hazard ratio of 1.61 (95% confidence interval 1.42–1.82) for death within 28 days of testing among individuals infected with lineage B.1.1.7; no significant differences in the increased hazard of death associated with lineage B.1.1.7 were found among individuals differing in age, sex, ethnicity, deprivation level, or place of residence. Both studies adjusted for varying COVID-19 mortality by geographical region and over time, correcting for potential biases due to differences in testing rates or differences in the availability of hospital services over time and space.
A Danish study found people infected by lineage B.1.1.7 to be 64% (32%–104%) more likely to get admitted to hospitals compared with people infected by another lineage.
Genetic sequencing of VOC-202012/01 has shown a Q27stop mutation which "truncates the ORF8 protein or renders it inactive". An earlier study of SARS-CoV-2 variants which deleted the ORF8 gene noted that they "have been associated to milder symptoms and better disease outcome". The study also noted that "SARS-CoV-2 ORF8 is an immunoglobulin (Ig)–like protein that modulates pathogenesis", that "SARS-CoV-2 ORF8 mediates major histocompatibility complex I (MHC-I) degradation", and that "SARS-CoV-2 ORF8 suppresses type I interferon (IFN)–mediated antiviral response".
Referring to amino-acid position 501 inside the spike protein, Chand et al. concluded that "it is possible that variants at this position affect the efficacy of neutralisation of virus", but noted that "here is currently no neutralisation data on N501Y available from polyclonal sera from natural infection". The HV 69–70 deletion has, however, been discovered "in viruses that eluded the immune response in some immunocompromised patients", and has also been found "in association with other RBD changes".
Epidemiology
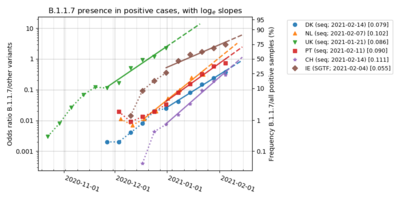

Cases of the Alpha variant (lineage B.1.1.7) were estimated to be under-reported by most countries as the most commonly used tests do not distinguish between this variant and other SARS-CoV-2 variants, and as many SARS-CoV-2 infections are not detected at all. RNA sequencing is required for detection of this variant, although RT-PCR test for specific variants can be used as a proxy test for Alpha — or as a supplementing first-screening test before conducting the whole-genome sequencing.
As of 23 March, the Alpha variant had been detected in 125 out of 241 sovereign states and dependencies (or 104 out of 194 WHO member countries), of which some had reported existence of community transmission while others so far only found travel related cases. As of 16 March, it had become the dominant COVID-19 variant for 21 countries: United Kingdom (week 52), Ireland (week 2), Bulgaria (week 4), Slovakia (week 5), Israel (week 5), Luxembourg (week 5), Portugal (week 6), Denmark (week 7), Netherlands (week 7), Norway (week 7), Italy (week 7), Belgium (week 7), France (week 8), Austria (week 8), Switzerland (week 8), Liechtenstein (week 9), Germany (week 9), Sweden (week 9), Spain (week 9), Malta (week 10) and Poland (week 11). The emergence and the fast spreading of the new variant has been detected in Lebanon and a relationship noted between SARS-CoV-2 transmission intensity and the frequency of the new variant during the first twelve days of January. By February, Alpha became the dominant variant in Lebanon.
Spread in UK
The first case was likely in mid-September 2020 in London or Kent, United Kingdom. The variant was sequenced in September. As of 13 December 2020, 1,108 cases with this variant had been identified in the UK in nearly 60 different local authorities. These cases were predominantly in the south east of England. The variant has also been identified in Wales and Scotland. By November, around a quarter of cases in the COVID-19 pandemic in London were being caused by the new variant, and by December, that was a third. In mid-December, it was estimated that almost 60 percent of cases in London involved Alpha. By 25 January 2021, the number of confirmed and probable UK cases had grown to 28,122.
Spread in Europe
The variant became dominant for:
- South East England in week 48, the last week of November 2020.
- England in week 51 of 2020.
- United Kingdom in week 52 of 2020.
- Scotland and Northern Ireland in week 1 of 2021.
- Wales in week 2 of 2021.
- Ireland in week 2 of 2021.
In Bulgaria, genome sequencing found the variant to be dominant with 52.1% in week 4, followed by 73.4% in week 9.

Also in Slovakia, a RT-PCR Multiplex DX test capable of detecting the 2 deletions specific for lineage B.1.1.7 (ΔH69/ΔV70 and ΔY144), first found the variant nationwide in 74% of cases on 3 February (week 5), followed by 72% of cases on 15 February (week 7), and it then grew to 90% of cases on 3 March (week 9). The same test found earlier on 8 January prevalence of the variant at a rate of 36% in the Michalovce District and 29% in Nitra.
In Israel, the variant was first time detected by genome sequencing 23 December 2020. Leumit Health Care Services however analysed with the proxy test RT-PCR (SGTF) and found the variant at a rate of 3‑4% on 15 December. The national Ministry of Health estimated based on genome sequencing, that the prevalence of the variant became dominant (70%) on 6 February followed by 90% on 16 February.
In Luxembourg, a weekly genome sequencing revealed that the variant grew from 0.3% (week 51) to be dominant with a share of 53.0% in week 5, although results might not be fully representative due to the fact that no correction occurred from potential targeting bias from contact tracing, airport travellers and local outbreaks. Genome sequencing of a population representative randomized test pool - with no target bias - was conducted since week 6, and it confirmed the dominant status of the variant at a rate of 54.1% (week 6) growing to 62.8% (week 9), while the competing Beta variant (lineage B.1.351) was found to be at 18.5% in week 9.
In Denmark the variant grew from 0.3% (week 46 of 2020) to become dominant with 65.9% (week 7 of 2021), and it grew further to 92.7% (week 10); with the regional prevalence ranging from 87.3% in the North Jutland Region to 96.1% in the Central Denmark Region. The observed growth of the relative variant share, was in full accordance with the earlier modelled forecast, that had predicted dominance (over 50%) around mid-February and a prevalence of around 80% of the total circulating variants by early March. In comparison, the genome sequencing only found the competing Beta variant in 0.4% of the positive cases (9 times out of 2315 tests) in week 10.
In the Netherlands, a randomly conducted genome sequencing found that the variant grew from 1.3% of cases in week 49 to a dominant share of 61.3% in week 7, followed by 82.0% in week 9; while the competing Beta variant in comparison was found to be at 3.0% in week 9. In Amsterdam, the Alpha variant (lineage B.1.1.7) grew from 5.2% (week 52) to 54.5% (week 6).
In Norway, the variant was found by genome sequencing to grow from 5.7% (week 1) into dominance by a 58.4% (week 7), followed by 65.0% (week 8). Another large survey comprising results of both genome sequencing and PCR proxy tests, with a sample seize of more than 1000 tests per week (since week 4), at the same time found that the variant grew from 2.0% (week 48) into dominance by 60.0% (week 7), followed by 72.7% in week 10 - while only 2.2% of cases in comparison were found to be of the Beta variant. The variant regionally had its highest share in the county of Oslo and Viken, growing from 18% to 90% of analysed samples in Oslo from 20 January to 23 February (although with the data-corrected estimate a bit lower at 50-70% on 23 February); while growing from 21% to 80% of analysed samples in Viken from 25 January to 23 February (although with the data-corrected estimate a bit lower at 50% on 23 February). For the period 15 February to 14 March, the combined survey of genome sequencing and PCR proxy tests, also found the Alpha variant was at a dominant rate over 50% for 8 out of 11 regions, with its highest rate (82%) found for Oslo; while the region Nordland was different from all other regions by having only 6% cases of Alpha along with a dominant 88% of cases represented by the Beta variant.
In Portugal, the variant represented according to a national genome sequencing survey: 16.0% of the Covid-19 infections during 10–19 January (week 2), followed by a dominant 58.2% in week 6. A national RT-PCR proxy test based on SGTF and SGTL observations, found the variant at a rate of 33.5% in week 4, but observed afterwards a decelerating pace for the weekly rise of the variant share (reason unknown), and according to this study it only became dominant by 50.5% (91.8% of 55.0% SGTFL) in week 8, followed by 64.3% (91.8% of 70% SGTFL) in week 10.
In Italy, the Alpha variant accounted for 17.8% of cases nationwide on 4–5 February (week 5), followed by 54.0% on 18 February (week 7). The regional prevalence for week 7 ranged from 0% in Aosta Valley (although only one sample was tested) to 93.3% in Molise. In week 7, the competing Gamma variant had a prevalence of 4.3% (ranging between 0%-36.2% regionally) and the South African variant a prevalence of 0.4% (ranging between 0%-2.9% regionally).
In Switzerland, a nationwide weekly genome sequencing survey found that the Alpha variant grew from 0.05% (week 51) to a dominant 58.2% of cases in week 8, followed by 71.1% in week 9. This was in full accordance with a model from 9 February that had forecast dominance around mid-February. In comparison, the competing Beta variant was only found nationwide in 1.0% of the positive cases in week 9.
In Belgium, genome sequencing of samples selected randomly after excluding all samples from active targeted testing related to local outbreaks or travels (creating a statistical representative national sample with a seize equal to 4.4% of all COVID-19 positive tests), found that the Alpha variant share grew from 7.1% in week 1 to a dominant 51.5% in week 7, followed by 79.3% in week 10. The variant was first time detected by targeted genome sequencing in week 49, but due to a small sample seize (not being random, and less than 100 tests per week) then no reliable variant share data could be determined before week 1. The proxy test for the variant (RT-PCR SGTF) was also conducted for a sample seize equal to 25.8% of all COVID-19 positive tests, and found a dominant 54.8% SGTF rate for week 10. In comparison, the competing Beta variant was found to be at 3.6% and the Gamma variant had a prevalence of 1.8% in week 10.
In France, scientists accurately forecast the Alpha variant (VOC-202012/01) would likely become dominant nationwide around week 8–11 of 2021. A nationwide survey of randomly selected positive COVID-19 samples first analysed by a RT-PCR screening test and subsequently confirmed by genome sequencing, revealed that the variant grew from a share of 3.3% (388/11916) on 7–8 January (week 1) to 13.3% (475/3561) on 27 January (week 4), followed by 44.3% (273/615) on 16 February (week 7). In week 8, the variant was found to have a dominant share of 56.4% (758/1345) according to the interpretable results of a weekly genome sequencing survey comprising 0.9% of all COVID-19 positive tests, or 59.5% according to a variant-specific RT-PCR survey testing 54% of all the COVID-19 positive tests. In week 10, the variant was found to have a share of 71.9% according to a variant-specific RT-PCR survey testing 56.9% of all the COVID-19 positive tests. The spread of the variant differed regionally for the 96 departments located in Metropolitan France for week 10, with 91 departments over 50% and 5 departments with 30%-50% (of which the Moselle department in particular was notable due to finding a high 38.3% rate of the competing Beta variant, that - despite having declined from a dominant 54.4% value in week 8 - still was significantly above the 5.0% national average for this specific variant).
In Austria, Agentur für Gesundheit und Ernährungssicherheit (AGES) collected data from N501Y RT-PCR specific tests combined with subsequent genome sequencing analysis, and found that the variant grew from 7.2% (week 1) to a dominant 61.3% (week 8), followed by 61.2% in week 9 and 48.3% in week 10. If all N501Y positive tests had been analysed further by genome sequencing, then these listed shares could have been even higher, for example they could have been as mcuh as: 2.4% higher for week 1, 23.0% higher for week 7, 4.0% higher for week 8, 6.8% higher for week 9 and 25.4% higher for week 10. The competing Beta variant was only found nationwide in 0.3% of the positive cases in week 10, and for the region Tyrol - where it had been most prevalent - its share declined from 24.5% in week 4 to just 1.9% in week 10. Regionally Alpha was found to be dominant with over 50% for 7 out of 9 regions, with the only two exceptions being Tyrol and Vorarlberg.
In Germany, the largest and probably most representative national survey published by the Robert Koch Institute (entitled RKI-Testzahlerfassung), determined the share of circulating COVID-19 variants for the latest week by analysing 53,272 COVID-19 positive samples either by genome sequencing or RT-PCR proxy tests, with data collected on a voluntary basis from 84 laboratories from the university / research / clinical / outpatient sector - spread evenly across the nation. The survey did not utilize data weights or data selection criteria to ensure existence of geographical representativity, but might still be regarded as somewhat representative for the national average due to its big sample seize. According to the RKI-Testzahlerfassung survey, the variant grew from a share of 2.0% (week 2) to a dominant share of 54.5% (week 9), followed by 63.5% in week 10. In comparison, the competing Beta variant was only found nationwide in 0.9% of the positive cases in week 10.
In Malta, the variant was first time detected by genome sequencing on 30 December 2020, and represented 8% of the positive cases in week 7. A new RT-PCR variant specific test was introduced for the surveillance, where the first results reported on 10 March revealed the variant now represented 61% of cases nationwide.
In Sweden, the national authorities initially expected the variant would become dominant around week 12–14 under the assumption of 50% increased transmissibility compared to the original virus. In average, the variant share was found growing from 10.8% (week 4) to 36.9% (week 7) across five of its southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro). For week 7, the share of the variant was for the first time also calculated to 30.4% as the overall average for 19 out of 21 Swedish regions (ranging from 3.3% in Blekinge to 45% in Gävleborg). For week 9, the share of the variant was calculated to a dominant 56.4% as the overall average for the 19 regions (ranging from 16% in Kronoberg to 72% in Gävleborg). For week 10, the share of the variant was calculated to have increased further to 71.3% as the overall average for the 19 regions (ranging from 40% in Kronoberg to 84% in Jönköping). Although no geographical weights were applied to ensure geographical representativity for the calculated average for the 19 regions, the overall sample seize of 12,417 variant tests represented 43.1% of all COVID-19 positive PCR-tests for week 10, inferring that the result of the survey might be close to represent the actual true average for the nation as a whole.
In the microstate Liechtenstein, the first case of Alpha was detected on 19 December 2020. During the entire pandemic, 67 VOC-N501Y PCR-positive cases were detected comprising 58 cases of Alpha, 1 of Beta (detected 1 February) and 8 un-identified N501Y cases (from 10 Dec. to 22 Feb.), as of 18 March 2021. The weekly average of each days calculated 7-day rolling average for the share of all detected VOC-N501Y cases (which is a good proxy for Alpha in Liechtenstein), was found to be 10.9% in week 5, followed by a dominant 52.2% in week 9 (where 8 out of 15 tests were found to be Alpha); and grew further to 73.4% in week 10 (where 6 out of 9 tests were found to be Alpha).
In Spain, the variant share was estimated nationwide to be 5%-10% of cases on 29 January, followed by 20%-25% of cases on 18 February and 25%-30% of cases on 22 February, and estimated to be dominant with over 50% as of 3 March (week 9). As of week 10, the prevalence of the variant ranged from 18.3% to 97.0% for the 17 regions, with all but two regions having a dominant rate above 50%:
- No data reported by Extremadura.
- 18.3% for week 10 in Aragon.
- 27.0% for week 7 in Canary Islands.
- 51.3% for week 8 in Castile and León.
- 52.2% for week 10 in Valencia.
- 52.8% for week 10 in Castilla–La Mancha.
- 53.3% for week 4 in Galicia.
- 59.5% for week 9 in La Rioja.
- 59.7% for week 10 in Madrid.
- 61.6% for week 11 in Andalusia.
- 76.1% for week 10 in Balearic Islands.
- 76.7% for week 10 in Murcia.
- 77.4% for week 10 in Basque Country.
- 77.8% for week 10 in Navarre.
- 83.1% for week 10 in Cantabria.
- 84.6% for week 9 in Catalonia.
- 97.0% for week 10 in Asturias.
In Poland, a national survey among infected teachers led to experts estimating that the variant share was between 5% and 10% nationwide as of 11 February, while ECDC reported it to be 9% as of 15 February. According to Health minister Adam Niedzielski, the variant was found at a rate of 5% in the first studies from the second half of January, and then it increased by ten percentage points every ten days, until it became dominant nationwide by a 52% rate on 16 March (week 11). Regionally, the variant had already exceeded 70% for Warmian-Masurian and Pomeranian on 22–23 February (week 8), and even reached 90% for the Greater Poland Voivodeship on 17 March (week 11).
In Finland, no statistical representative national survey had been conducted as of February 2021, as the national genome sequencing mainly targeted further analysis of COVID-19 positive samples from travellers and local outbreak clusters. Helsinky University Hospital (HUS), operating in the Helsinki and Uusimaa Hospital District, found the variant in 10% of all samples collected randomly during a few days ahead of 14 February in the capitol region (also known as the Helsinky constituency and Uusimaa constituency). For the capitol region, the variant was modelled to become dominant (over 50%) in the second half of March (week 11-13).
In Iceland, the national authorities implemented a strict 3 test and quarantine regime for all persons entering the country from abroad, that so far successfully managed to prevent new infectious VOCs from gaining a foothold in the country. As of 4 March, a total of 90 travellers had tested positive for the Alpha variant at Iceland's borders and the 20 additional domestic cases were all closely connected to the border cases, with no cases related to community transmission.
Spread in North America
In the United States, the variant first appeared late November 2020, grew from 1.2% in late January and became predominant around the end of March.The spread of the variant was primarily concentrated in the upper Midwest of the country, especially the states of Michigan and Minnesota, but it failed to spread in high concentrations to other parts of the country.
In Canada, the variant first appeared in Ontario late December 2020. By 13 February, it had spread to all ten provinces. Testing and confirmation of the Alpha variant (lineage B.1.1.7) in COVID-19-positive samples has been inconsistent across the country. On 3 February, the province of Alberta was the first to screen all COVID-19 positive samples for variants of concern. As of 23 March, the Alpha variant had been detected in 5812 cases, and was most prevalent in the province of Alberta. In Ontario, a combined RT-PCR (N501Y) and genome sequencing study found that all VOCs represented 4.4% of all COVID-19 positives on 20 January (week 3), and that Alpha comprised 99% of all those VOCs. Public Health Ontario laboratories found the variant in close to 7% of all COVID-19 positives in week 5, representing 97% (309/319) of all detected VOCs. Alpha became the dominant variant on or around 16 March; 53% of all positive cases were VOCs, and it was presumed 97% of VOCs were Alpha. In Quebec, where the variant was also widespread, it was expected to become dominant by late March or in April.
Development
| Development of the Alpha variant (lineage B.1.1.7) (share of analysed SARS-CoV-2–positive tests in a given week) | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | Test | 2020 Week 42 | Week 43 | Week 44 | Week 45 | Week 46 | Week 47 | Week 48 | Week 49 | Week 50 | Week 51 | Week 52 | Week 53 | 2021 Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 |
| United Kingdom | Seq. | 0.05% | 0.35% | 1.0% | 2.7% | 6.3% | 10.2% | 10.2% | 13.9% | 32.9% | 45.7% | 51.3% | 70.6% | 74.1% | 78.6% | 86.0% | 89.1% | – | – | – | – | – | – |
| England | SGTF*Seq.% Seq. |
0.07% (0.02%) |
0.4% (0.5%) |
0.9% (1.2%) |
2.9% (1.5%) |
5.5% (5.3%) |
9.1% (7.8%) |
18.0% (11.4%) |
32.2% (22.1%) |
51.8% (38.1%) |
63.7% (61.8%) |
72.4% (62.1%) |
77.9% (78.4%) |
82.1% (76.7%) |
87.5% (79.7%) |
90.6% (–) |
93.8% (–) |
95.7% (–) |
97.4% (–) |
98.1% (–) |
98.6% (–) |
98.7% (–) |
– (–) |
| Northern Ireland | SGTF rawdata SGTF model |
N/A | N/A | N/A | N/A | N/A | 17.9% (m:14.0%) |
7.4% (m:14.9%) |
37.5% (m:16.3%) |
23.0% (m:18.5%) |
50.0% (m:36.1%) |
25.0% (m:42.1%) |
58.8% (m:44.7%) |
45.2% (m:52.7%) |
71.4% (m:67.0%) |
78.2% (m:74.0%) |
72.4% (m:81.4%) |
95.7% (m:86.5%) |
88.9% (m:90.5%) |
92.2% (m:92.4%) |
100.0% (m:93.0%) |
86.2% (m:92.6%) |
100.0% (m:91.1%) |
| Scotland | SGTF rawdata SGTF model |
N/A | N/A | N/A | N/A | N/A | 9.5% (m:5.8%) |
9.7% (m:6.7%) |
10.4% (m:7.7%) |
11.7% (m:10.2%) |
51.4% (m:26.2%) |
40.0% (m:39.5%) |
35.8% (m:43.9%) |
64.1% (m:49.8%) |
67.2% (m:64.2%) |
63.3% (m:65.0%) |
69.2% (m:70.0%) |
70.6% (m:77.3%) |
93.9% (m:84.7%) |
82.0% (m:90.0%) |
100.0% (m:93.7%) |
93.0% (m:95.8%) |
100.0% (m:96.9%) |
| Wales | SGTF rawdata SGTF model |
N/A | N/A | N/A | N/A | N/A | 27.9% (m:18.1%) |
8.3% (m:16.0%) |
10.7% (m:13.3%) |
26.3% (m:12.1%) |
13.4% (m:15.0%) |
15.9% (m:14.7%) |
19.8% (m:20.4%) |
54.4% (m:34.8%) |
61.5% (m:59.6%) |
69.0% (m:65.0%) |
68.1% (m:68.7%) |
77.0% (m:73.4%) |
71.8% (m:76.6%) |
75.8% (m:78.5%) |
87.9% (m:78.9%) |
80.0% (m:77.2%) |
69.9% (m:73.3%) |
| Ireland | SGTF rawdata | – | – | – | 1.9% (few data) |
0.0% (few data) |
0.0% (few data) |
6.1% (few data) |
2.2% (few data) |
1.6% (few data) |
7.5% | 16.3% | 26.2% | 46.3% | 57.7% | 69.5% | 75.0% | 90.1% | 88.6% | 90.8% | – | – | – |
| Bulgaria | Seq. | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 52.1% | – | 70.8% | – | – | 73.4% | – |
| Israel | SGTF w.51+1 Seq. after w.1 |
– | – | – | – | – | – | – | – | – | 3–4% (Dec.15) |
– | – | 10–20% (Jan.5) |
10–20% (Jan.11) |
30–40% (Jan.19) |
40–50% (Jan.25) |
~70% (Feb.2) |
~80% (Feb.9) |
~90% (Feb.16) |
– | – | – |
| Slovakia | PCR (SGTF+ΔY144) | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ~74% (Feb.3) |
– | ~72% (Feb.15) |
– | ~90% (Mar.3) |
– |
| Luxembourg | Seq. | – | – | – | – | – | – | – | – | – | 0.3% | 0.9% | 4.2% | 8.7% | 15.5% | 16.9% | 36.2% | 53.0% | 54.1% | 53.8% | 63.7% | 62.8% | – |
| Denmark | Seq. | – | – | – | – | 0.3% | 0.2% | 0.4% | 0.4% | 0.4% | 0.8% | 1.8% | 1.9% | 3.5% | 7.0% | 13.1% | 19.6% | 29.6% | 47.0% | 65.9% | 75.3% | 85.1% | 92.7% |
| Netherlands | Seq. | – | – | – | – | – | – | – | 1.3% | 0.8% | 0.5% | 2.1% | 4.4% | 9.6% | 15.9% | 22.7% | 24.7% | 31.0.% | 40.3% | 61.3% | 75.2% | 82.0% | – |
| Norway | PCR proxy and Seq. Seq. |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
0.0% (–) |
2.0% (–) |
0.0% (0.0%) |
4.9% (0.0%) |
8.9% (7.2%) |
6.0% (2.9%) |
14.9% (1.3%) |
18.9% (5.7%) |
19.5% (11.6%) |
15.2% (9.3%) |
19.8% (20.1%) |
34.1% (30.3%) |
43.6% (40.8%) |
60.0% (58.4%) |
71.8% (65.0%) |
76.7% (–) |
72.7% (–) |
| Italy | Seq.% | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 17.8% | – | 54.0% | – | – | – |
| France | PCR proxy Seq. SGTF*Seq.% |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (3.3%) |
– (–) (–) |
– (–) (–) |
– (–) (13.3%) |
– (–) (–) |
36.1% (–) (–) |
49.3% (45.8%) (44.3%) |
59.5% (56.4%) (–) |
65.8% (69.0%) (–) |
71.9% (–) (–) |
| Switzerland | Seq. | – | – | – | – | – | – | – | – | – | 0.05% | 0.4% | 0.8% | 2.6% | 4.9% | 9.2% | 16.5% | 26.0% | 32.4% | 47.0% | 58.2% | 71.1% | – |
| Austria | PCR(N501Y)*Seq.% Seq.B117+unseq.N501Y |
– | – | – | – | – | – | – | – | – | – | – | – | 7.2% (9.6%) |
17.4% (23.8%) |
14.7% (19.6%) |
22.5% (28.6%) |
28.7% (37.2%) |
24.0% (45.4%) |
35.1% (58.1%) |
61.3% (65.3%) |
61.2% (68.0%) |
48.3% (73.7%) |
| Belgium | Seq. SGTF |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
0.0% (<5%) |
0.0% (<5%) |
0.0% (<5%) |
13.6% (<5%) |
0.0% (<5%) |
7.1% (7.2%) |
7.7% (6.9%) |
13.4% (16.6%) |
23.3% (19.5%) |
39.4% (22.4%) |
43.2% (27.9%) |
51.5% (34.4%) |
57.6% (45.8%) |
66.5% (46.3%) |
79.3% (54.8%) |
| Portugal | SGTFL*Seq.% | – | – | – | – | – | – | – | 1.7% | 0.8% | 1.2% | 1.7% | 2.9% | 6.8% | 12.3% | 22.7% | 33.5% | 39.2% | 40.9% | 45.7% | 50.5% | 56.0% | 64.3% |
| Sweden | Seq. all N501Y+A570D pos. tests | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 10.8% (av. for 5 out of 21 regions) |
15.1% (av. for 5 out of 21 regions) |
27.3% (av. for 5 out of 21 regions) |
30.4% (av. for 19 out of 21 regions) |
41.5% (av. for 19 out of 21 regions) |
56.4% (av. for 17 out of 21 regions) |
71.3% (av. for 18 out of 21 regions) |
| Germany | PCR proxy and Seq. Seq. |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (2.6%) |
2.0% (8.8%) |
3.6% (4.9%) |
4.7% (10.7%) |
7.2% (17.7%) |
17.6% (20.8%) |
25.9% (33.2%) |
40.0% (42.9%) |
54.5% (48.3%) |
63.5% (–) |
| Liechtenstein | PCR proxy: N501Y | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 10.9% | 28.5% | 32.8% | 47.0% | 52.2% | 73.4% |
| Spain | Estimated based on PCR proxy | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 5-10% (Jan.29) |
– | – | 20-25% (Feb.18) |
25-30% (Feb.22) |
>50% (Mar.3) |
– |
| United States | SGTF*Seq.% | – | – | – | – | – | – | – | – | – | 0.05% | 0.2% | 0.4% | 0.5% | 1.0% | 1.9% | 3.0% | 4.7% | 7.6% | 12.4% | 19.3% | 28.8% | 37.4% |
| California | SGTF*Seq.% | – | – | – | – | – | – | – | – | – | 0.3% | 0.3% | 0.7% | 1.1% | 1.3% | 2.0% | 1.9% | 4.2% | 5.9% | 13.1% | 16.6% | 18.8% | 28.0% |
| Florida | SGTF*Seq.% | – | – | – | – | – | – | – | – | – | 0.2% | 0.5% | 0.9% | 1.0% | 2.3% | 4.6% | 8.1% | 10.7% | 14.5% | 21.0% | 28.3% | 39.8% | 47.0% |
| Other countries without weekly/periodic data, but with a variant share above 5%: 00 Malta (61%), and Poland (52%). | |||||||||||||||||||||||
The following additional countries did not report variant shares, but are likely to have a significant share present due to their finding of more than 50 cases confirmed by whole genome sequencing per 13 March 2021:
- Turkey (486 cases)
- Finland (268 cases)
- Australia (140 cases)
- Chile (124 cases)
- Croatia (121 cases)
- Nigeria (113 cases)
- South Korea (90 cases)
- Slovenia (87 cases)
- Latvia (83 cases)
- India (81 cases)
- Romania (76 cases)
- Ghana (67 cases)
- Singapore (66 cases)
- New Zealand (63 cases)
- North Macedonia (53 cases)
- Brazil (53 cases)
- Taiwan (32 cases)
The GISAID database of all sequenced COVID-19 genomes, calculates for each country for the past four weeks an average "Relative Variant Genome Frequency" for submitted samples. Those observed frequencies are however subject to sampling and reporting biases, and do not represent exact variant share prevalence due to absence of statistical representativity.
Countries reporting a first case
December 2020
Cases of the variant began to be reported globally during December, being reported in Denmark, Belgium, the Netherlands, Australia and Italy. Shortly after, several other countries confirmed their first cases, the first of whom were found in Iceland and Gibraltar, then Singapore, Israel and Northern Ireland on 23 December, Germany and Switzerland on 24 December, and the Republic of Ireland and Japan confirmed on 25 December.
The first cases in Canada, France, Lebanon, Spain and Sweden were reported on 26 December. Jordan, Norway, and Portugal reported their first case on 27 December, Finland and South Korea reported their first cases on 28 December, and Chile, India, Pakistan and the United Arab Emirates reported their first cases on 29 December. The first case of new variant in Malta and Taiwan are reported on 30 December. China and Brazil reported their first cases of the new variant on 31 December. The United Kingdom and Denmark are sequencing their SARS-CoV-2 cases at considerably higher rates than most others, and it was considered likely that additional countries would detect the variant later.
The United States reported a case in Colorado with no travel history on 29 December, the sample was taken on 24 December. On 6 January 2021, the US Centers for Disease Control and Prevention announced that it had found at least 52 confirmed cases in California, Florida, Colorado, Georgia, and New York. In the following days, more cases of the variant were reported in other states, leading former CDC director Tom Frieden to express his concerns that the U.S. will soon face "close to a worst-case scenario".
January 2021
Turkey detected its first cases in 15 people from England on 1 January 2021. It was reported on 1 January that Denmark had found a total of 86 cases of the variant, equalling an overall frequency of less than 1% of the sequenced cases in the period from its first detection in the country in mid-November to the end of December; this had increased to 1.6% of sequenced tests in the period from mid-November to week two of 2021, with 7% of sequenced tests in this week alone being of Alpha variant (lineage B.1.1.7). Luxembourg and Vietnam reported their first case of this variant on 2 January 2021.
On 3 January 2021, Greece and Jamaica detected their first four cases of this variant and Cyprus announced that it had detected Alpha (VOC 202012/01) in 12 samples. At the same time, New Zealand and Thailand reported their first cases of this variant, where the former reported six cases made up of five from the United Kingdom and one from South Africa, and the latter reported the cases from a family of four who had arrived from Kent. Georgia reported its first case and Austria reported their first four cases of this variant, along with one case of Beta variant, on 4 January.
On 5 January, Iran, Oman, and Slovakia reported their first cases of VOC-202012/01. On 8 January, Romania reported its first case of the variant, an adult woman from Giurgiu County who declared not having left the country recently. On 9 January, Peru confirmed its first case of the variant. Mexico and Russia reported their first case of this variant on 10 January, then Malaysia and Latvia on 11 January.
On 12 January, Ecuador confirmed its first case of this variant. The Philippines and Hungary both detected the presence of the variant on 13 January. The Gambia recorded first cases of the variant on 14 January with it being the first confirmation of the variant's presence in Africa. On 15 January, the Dominican Republic confirmed its first case of the new variant and Argentina confirmed its first case of the variant on 16 January. Czech Republic and Morocco reported their first cases on 18 January while Ghana and Kuwait confirmed their first cases on 19 January. Nigeria confirmed its first case on 25 January. On 28 January, Senegal detected its first case of the variant.
In early January, an outbreak linked to a primary school led to the detection of at least 30 cases of the new variant in the Bergschenhoek area of the Netherlands, signifying local transmission.
On 24 January, a person travelling from Africa to Faroe Islands tested positive upon arrival to the islands, and went directly into quarantine.
On 28 January, North Macedonia confirmed the variant was detected in a 46-year-old man, who had already recovered.
February 2021
On 1 February, Lithuania confirmed the first cases of the new lineage. On 4 February, health authorities in Uruguay announced the first case of the variant in the country. The case was detected in a person who entered the country on 20 December 2020 and has been in quarantine ever since. On 10 February, the Health Ministry of Croatia confirmed that out of 61 sequenced samples since 20 January, the variant was detected in 3 samples: a male 50 year old and 3.5 year old from Zagreb, and a male 34-year-old from Brod-Posavina County. On 12 February, the variant was detected from four areas in Sri Lanka, and the Canadian province of Newfoundland and Labrador confirmed an outbreak of the variant.
March 2021
On 2 March, Indonesia reports its first cases of the variant in two migrant workers returning from Saudi Arabia. On the same day, Tunisia reported their first cases of the variant. The presence of the variant in Ivory Coast was confirmed on 25 March.
N501Y mutation elsewhere
The N501Y mutation arose independently multiple times in different locations:
- In April 2020, it was seen for the first time in a few isolated sequences in Brazil.
- In June 2020, the mutation appeared in an Australian lineage.
- In July 2020, (according to Dr Julian Tang, University of Leicester) N501Y appeared in a lineage circulating in United States.
- In September 2020, it was found in a lineage in Wales, that evolved independently and was different from lineage B.1.1.7.
- In September 2020, the much more transmissible and widespread lineage B.1.1.7 (501Y.V1) was detected for the first time in Kent (England).
- In October 2020, another highly transmissible variant of concern termed lineage B.1.351 (501Y.V2, Beta variant) was first time detected by genome sequencing in Nelson Mendela Bay (South Africa). Phylogeographic analysis suggests this lineage emerged already in July or August 2020.
- In December 2020, another highly transmissible variant termed lineage P.1 (501Y.V3, Gamma variant) was detected in Manaus (Brazil).
Statistics
Table
| Country | Confirmed cases (GISAID) as of 16 January 2024 |
Last Case Reported |
|---|---|---|
| 268,555 | 27 June 2021 | |
| 250,995 | 6 July 2021 | |
| 219,985 | 29 June 2021 | |
| 102,642 | 28 June 2021 | |
| 62,570 | 29 June 2021 | |
| 58,838 | 21 June 2021 | |
| 32,782 | 28 June 2021 | |
| 28,773 | 25 June 2021 | |
| 32,899 | 1 July 2021 | |
| 25,420 | 30 June 2021 | |
| 21,726 | 26 June 2021 | |
| 22,258 | 5 July 2021 | |
| 20,724 | 1 July 2021 | |
| 14,804 | 21 June 2021 | |
| 15,798 | 23 June 2021 | |
| 9,334 | 11 June 2021 | |
| 7,988 | 24 June 2021 | |
| 8,526 | 23 June 2021 | |
| 8,366 | 20 June 2021 | |
| 7,953 | 25 May 2021 | |
| 5,481 | 7 June 2021 | |
| 5,015 | 23 June 2021 | |
| 4,480 | 19 June 2021 | |
| 4,898 | 25 May 2021 | |
| 4,350 | 11 June 2021 | |
| 3,836 | 20 June 2021 | |
| 4,299 | 24 June 2021 | |
| 3,361 | 1 June 2021 | |
| 3,135 | 28 May 2021 | |
| 2,993 | 8 May 2021 | |
| 3,056 | 17 May 2021 | |
| 1,705 | 24 June 2021 | |
| 726 | 20 June 2021 | |
| 767 | 15 June 2021 | |
| 1,001 | 30 March 2021 | |
| 637 | 25 May 2021 | |
| 735 | 8 June 2021 | |
| 551 | 16 June 2021 | |
| 584 | 1 June 2021 | |
| 506 | 4 July 2021 | |
| 559 | 3 June 2021 | |
| 470 | 26 June 2021 | |
| 384 | 24 June 2021 | |
| 361 | 12 June 2021 | |
| 318 | 19 May 2021 | |
| 386 | 25 June 2021 | |
| 347 | 24 April 2021 | |
| 258 | 22 April 2021 | |
| 231 | 23 May 2021 | |
| 190 | 17 June 2021 | |
| 179 | 28 May 2021 | |
| 183 | 15 June 2021 | |
| 162 | 27 April 2021 | |
| 152 | 14 June 2021 | |
| 148 | 11 June 2021 | |
| 162 | 12 June 2021 | |
| 140 | 19 March 2021 | |
| 165 | 17 June 2021 | |
| 131 | 8 March 2021 | |
| 139 | 7 May 2021 | |
| 167 | 17 June 2021 | |
| 159 | 7 June 2021 | |
| 110 | 10 March 2021 | |
| 106 | 22 April 2021 | |
| 129 | 24 May 2021 | |
| 93 | 8 June 2021 | |
| 99 | 25 May 2021 | |
| 90 | 4 May 2021 | |
| 68 | 21 June 2021 | |
| 48 | 9 March 2021 | |
| 87 | 8 June 2021 | |
| 80 | 8 June 2021 | |
| 79 | 23 April 2021 | |
| 72 | 25 March 2021 | |
| 63 | 28 June 2021 | |
| 70 | 18 March 2021 | |
| 52 | 8 May 2021 | |
| 54 | 14 June 2021 | |
| 59 | 22 May 2021 | |
| 39 | 11 May 2021 | |
| 44 | 5 May 2021 | |
| 31 | 5 February 2021 | |
| 59 | 29 May 2021 | |
| 62 | 31 March 2021 | |
| 29 | 26 February 2021 | |
| 28 | 11 March 2021 | |
| 22 | 9 April 2021 | |
| 25 | 6 May 2021 | |
| 66 | 6 April 2021 | |
| 45 | 11 March 2021 | |
| 21 | 5 June 2021 | |
| 154 | 25 May 2021 | |
| 21 | 25 February 2021 | |
| 19 | 29 December 2020 | |
| 26 | 21 May 2021 | |
| 20 | 5 January 2021 | |
| 28 | 29 April 2021 | |
| 35 | 30 April 2021 | |
| 17 | 15 June 2021 | |
| 767 | 3 May 2021 | |
| 15 | 9 March 2021 | |
| 152 | 12 February 2021 | |
| 15 | 14 May 2021 | |
| 7 | 18 March 2021 | |
| 11 | 17 April 2021 | |
| 7 | 17 May 2021 | |
| 25 | 30 March 2021 | |
| 31 | 4 June 2021 | |
| 11 | 2 June 2021 | |
| 10 | 27 January 2021 | |
| 10 | 19 April 2021 | |
| 9 | 4 May 2021 | |
| 16 | 28 March 2021 | |
| 16 | 14 April 2021 | |
| 17 | 7 April 2021 | |
| 14 | 16 May 2021 | |
| 7 | 21 January 2021 | |
| 6 | 21 May 2021 | |
| 12 | 6 May 2021 | |
| 10 | 15 May 2021 | |
| 5 | 22 March 2021 | |
| 16 | 1 April 2021 | |
| 4 | 8 April 2021 | |
| 4 | 11 June 2021 | |
| 3 | 7 February 2021 | |
| 3 | 24 February 2021 | |
| 3 | 3 February 2021 | |
| 3 | 4 February 2021 | |
| 22 | 6 January 2021 | |
| 148 | 10 June 2021 | |
| 16 | 3 March 2021 | |
| 3 | 17 April 2021 | |
| 29 | 5 March 2021 | |
| 6 | 8 March 2021 | |
| 2 | 23 March 2021 | |
| 2 | 5 January 2021 | |
| 2 | 17 March 2021 | |
| 2 | 4 May 2021 | |
| 3 | 8 April 2021 | |
| 2 | 8 March 2021 | |
| 2 | 21 May 2021 | |
| 2 | 5 February 2021 | |
| 2 | 28 May 2021 | |
| 6 | 5 March 2021 | |
| 2 | 11 February 2021 | |
| 1 | 20 January 2021 | |
| 1 | 19 April 2021 | |
| 1 | 9 February 2021 | |
| 17 | 7 April 2021 | |
| 1 | 19 May 2021 | |
| 1 | 23 April 2021 | |
| 1 | 10 January 2021 | |
| 1 | 21 April 2021 | |
| 1 | 29 April 2021 | |
| 30 | 22 December 2020 | |
| 4 | 15 April 2021 | |
| 1 | 9 January 2021 | |
| 1 | 5 June 2021 | |
| 1 | 3 April 2021 | |
| 1 | 27 May 2021 | |
| 1 | - | |
| 2 | 25 June 2021 | |
| 4 | 3 February 2021 | |
| 15 | 3 March 2021 | |
| 3 | March 2021 | |
| 4 | 29 March 2021 | |
| 3 | 7 May 2021 | |
| World (167 countries) | Total: 1,248,973 | Total as of 13 August 2021 |
Graphics
Confirmed cases by countries
- Note: The graphs presented here are only viewable by computers and some phones. If you cannot view it on your cell phone, switch to desktop mode from your browser.
- Data provided by various sources, such as; governmental, press or officials are updated every week since their last publication.
| Graphs are unavailable due to technical issues. Updates on reimplementing the Graph extension, which will be known as the Chart extension, can be found on Phabricator and on MediaWiki.org. |
| Graphs are unavailable due to technical issues. Updates on reimplementing the Graph extension, which will be known as the Chart extension, can be found on Phabricator and on MediaWiki.org. |
| Graphs are unavailable due to technical issues. Updates on reimplementing the Graph extension, which will be known as the Chart extension, can be found on Phabricator and on MediaWiki.org. |
| Graphs are unavailable due to technical issues. Updates on reimplementing the Graph extension, which will be known as the Chart extension, can be found on Phabricator and on MediaWiki.org. |
History
Detection

The Alpha variant (lineage B.1.1.7) was first detected in early December 2020 by analysing genome data with knowledge that the rates of infection in Kent were not falling despite national restrictions.
The two earliest genomes that belong to lineage B.1.1.7 were collected on 20 September 2020 in Kent and another on 21 September 2020 in Greater London. These sequences were submitted to the GISAID sequence database (sequence accessions EPI_ISL_601443 and EPI_ISL_581117, respectively).
Backwards tracing using genetic evidence suggests lineage B.1.1.7 emerged in September 2020 and then circulated at very low levels in the population until mid-November. The increase in cases linked to the variant first became apparent in late November when Public Health England (PHE) was investigating why infection rates in Kent were not falling despite national restrictions. PHE then discovered a cluster linked to this variant spreading rapidly into London and Essex.
Also important was the nature of the RT-PCR test used predominantly in the UK, Thermo Fisher Scientific's TaqPath COVID-19. The test matches RNA in three locations, and stopped working for the spike gene due to the HV 69–70 deletion—a deletion of the amino acids histidine and valine in positions 69 and 70, respectively—in the spike protein of lineage B.1.1.7. This made preliminary identification easier because it could be better suspected which cases were with lineage B.1.1.7 through genome sequencing.
It has been suggested that the variant may have originated in a chronically infected immunocompromised person, giving the virus a long time to replicate and evolve.
Control
In the presence of a more transmissible variant, stronger physical distancing and lockdown measures were opted for to avoid overwhelming the population due to its tendency to grow exponentially.
All countries of the United Kingdom were affected by domestic travel restrictions in reaction to the increased spread of the virus—at least partly attributed to Alpha—effective from 20 December 2020. During December 2020, an increasing number of countries around the world either announced temporary bans on, or were considering banning, passenger travel from the UK, and in several cases from other countries such as the Netherlands and Denmark. Some countries banned flights; others allowed only their nationals to enter, subject to a negative SARS-CoV-2 test. A WHO spokesperson said that, "cross Europe, where transmission is intense and widespread, countries need to redouble their control and prevention approaches". Most bans by EU countries were for 48 hours, pending an integrated political crisis response meeting of EU representatives on 21 December to evaluate the threat from the new variant and coordinate a joint response.
Many countries around the world imposed restrictions on passenger travel from the United Kingdom; neighbouring France also restricted manned goods vehicles (imposing a total ban before devising a testing protocol and permitting their passage once more). Some also applied restrictions on travel from other countries. As of 21 December 2020, at least 42 countries had restricted flights from the UK, and Japan was restricting entry of all foreign nationals after cases of the new variant were detected in the country.
The usefulness of travel bans has been contested as limited in cases where the variant has likely already arrived, especially if the estimated growth rate per week of the virus is higher locally.
Extinction
In October 2021, Dr Jenny Harries, chief executive of the UK Health and Security Agency stated that previous circulating variants such as Alpha had 'disappeared' and replaced by the Delta variant. In March 2022, the World Health Organization listed the Alpha, Beta and Gamma variants as previously circulating citing lack of any detected cases in the prior weeks and months.
See also
- Variants of SARS-CoV-2: Beta, Gamma, Delta, Epsilon, Zeta, Eta, Theta, Iota, Kappa, Lambda, Mu, Omicron
Notes
- Written as VUI 202012/01 (Variant Under Investigation, year 2020, month 12, variant 01) by GISAID and the ECDC.
- The difference between the two is explained by PHE:
SARS-CoV-2 variants, if considered to have concerning epidemiological, immunological, or pathogenic properties, are raised for formal investigation. At this point they are designated Variant Under Investigation (VUI) with a year, month, and number. Following a risk assessment with the relevant expert committee, they may be designated Variant of Concern (VOC).
- ^ SARS-CoV-2's S gene encodes its spike protein.
- An example of this is the delta (Δ)–PCR test, which in connection to SARS-CoV-2, has been used to detect the HV 69–70 deletion in variants with this mutation through what has been named "spike-gene target failure" (SGTF) or "spike-gene drop out" for the spike (S) gene in a subset of RT-PCR assays (e.g., TaqPath COVID-19 RT-PCR assay, ThermoFisher). Though existing in a few other variants of SARS-CoV-2, the HV 69–70 deletion in the spike protein is present in the vast majority of B.1.1.7 genomes, which enables the delta-PCR test to be used as a proxy test for the lineage—or as a supplementing first-screening test before conducting the whole-genome sequencing.
Another example of a RT-PCR test intended to detect specific variants is the one detecting all genomes with the N501Y mutation (e.g., Alpha, Gamma variant, and Beta variant), which is now also being used as a first-step screening tool ahead of genome sequencing by several laboratories/countries (e.g. by some parts of France).
A third and fourth RT-PCR test intended for detecting specific variants pre-screen the samples for variants respectively with the N501Y+A570D mutations (Alpha) and N501Y without the A570D mutation (Beta, Gamma, and other N501Y variants). - 104 WHO members had reported a detection of the Alpha variant (lineage B.1.1.7), as of 23 March 2021: Albania, Angola, Argentina, Australia, Austria, Azerbaijan, Bahrain, Bangladesh, Barbados, Belarus, Belgium, Belize, Bosnia and Herzegovina, Brazil, Brunei Darussalam, Bulgaria, Cabo Verde, Cambodia, Canada, Chile, China, Costa Rica, Croatia, Cyprus, Czech Republic, Democratic Republic of the Congo, Denmark, Dominican Republic, Ecuador, Estonia, Finland, France, Gambia, Georgia, Germany, Ghana, Greece, Hungary, Iceland, India, Indonesia, Iran, Iraq, Ireland, Israel, Italy, Jamaica, Japan, Jordan, Kenya, Kuwait, Latvia, Lebanon, Libya, Lithuania, Luxembourg, Malaysia, Malta, Mauritania, Mauritius, Mexico, Monaco, Montenegro, Morocco, Nepal, Netherlands, New Zealand, Nigeria, North Macedonia, Norway, Oman, Pakistan, Peru, Philippines, Poland, Portugal, Republic of Korea (South Korea), Republic of Moldova, Romania, Russia, Rwanda, Saint Lucia, Saudi Arabia, Senegal, Serbia, Singapore, Slovakia, Slovenia, South Africa, Spain, Sri Lanka, Sweden, Switzerland, Thailand, Trinidad and Tobago, Tunisia, Turkey, Ukraine, United Arab Emirates, United Kingdom, United States, Uruguay, Uzbekistan, Vietnam.
90 WHO members had not reported any detection of the Alpha variant (lineage B.1.1.7), as of 23 March 2021: Afghanistan, Algeria, Andorra, Antigua and Barbuda, Armenia, Bahamas, Benin, Bhutan, Bolivia, Botswana, Burkina Faso, Burundi, Cameroon, Central African Republic, Chad, Colombia, Comoros, Congo, Cook Islands, Côte d'Ivoire, Cuba, Democratic People's Republic of Korea (North Korea), Djibouti, Dominica, Egypt, El Salvador, Equatorial Guinea, Eritrea, Ethiopia, Fiji, Gabon, Grenada, Guatemala, Guinea-Bissau, Guyana, Haiti, Honduras, Guinea, Kazakhstan, Kiribati, Kyrgyzstan, Lao People's Democratic Republic, Lesotho, Liberia, Madagascar, Malawi, Maldives, Mali, Marshall Islands, Micronesia, Mongolia, Mozambique, Myanmar, Namibia, Nauru, Nicaragua, Niger, Niue, Palau, Panama, Papua New Guinea, Paraguay, Qatar, Saint Kitts and Nevis, Saint Vincent and the Grenadines, Samoa, San Marino, Sao Tome and Principe, Seychelles, Sierra Leone, Solomon Islands, Somalia, South Sudan, Sudan, Suriname, Swaziland, Syrian Arab Republic, Tajikistan, Timor-Leste, Togo, Tonga, Turkmenistan, Tuvalu, Uganda, United Republic of Tanzania, Vanuatu, Venezuela, Yemen, Zambia, Zimbabwe. - ^ The weekly UK infection survey lists for each country in Great Britain a set of raw data and average-smoothed modelled data from SGTF analysed PCR tests collected from private households (excl. tests from hospitals, care centres and public institutions). Raw data as well as model data for Wales, Northern Ireland and Scotland must be treated with caution due to a small number of collected samples, resulting in great data uncertainty.
- All PCR tests were analysed for 3 genes present in the coronavirus: N protein, S protein and ORF1ab (see Table 6A in the Infection Survey). Each PCR test can have any one, any two or all three genes detected. Coronavirus positives are those where one or more of these genes is detected in the swab (other than tests that are only positive on the S-gene which is not considered a reliable indicator of the virus if found on its own). The new B.1.1.7 variant of COVID-19 has genetic changes in the S gene, that results in the S-gene no longer being detected in the current test, meaning that it will only be positive on the ORF1ab and the N gene. The survey uses the terms "New UK variant compatible" for ORF1ab + N protein gene positives, "not compatible with new UK variant" for ORF1ab + N protein + S protein gene positive, and "virus too low to be identifiable" for all other gene patterns (a reasonable definition given that all samples taken from the first phase of the COVID-19 disease where virus exist by identifiable quantity, either will be positive by "ORF1ab+N" or "ORF1ab+N+S"). However, further uncertainty exists given that not all "New UK variant compatible" SGTF cases (positive on ORF1ab and N-genes, but not the S-gene) will be the new UK variant - due to some other competing variants also delivering this same test pattern; and prior to mid-November 2020 the data should not be read as being an indicator of the variant at all.
- The weekly B.1.1.7 variant share raw data is calculated from Table 6A, by dividing the "ORF1ab+N" percentage with the percentage sum of "ORF1ab+N" and "ORF1ab+N+S". Table 6C utilized the raw data from table 6A as input for calculation of some modelled (average-smoothed and weighted) daily estimated figures for the respective percentage of the population being coronavirus positive by either the "New UK variant compatible virus", or a "Not compatible with new UK variant virus" or a "virus too low to be identifiable", with data from earlier dates in 2020 also being available when downloading the published earlier editions of the Infection Survey. The modelled weekly average value for the Alpha variant, noted in this table as the value (m: %), is calculated as the average for the listed seven days in each week of "New UK variant compatible percentage" divided by the sum of "New UK variant compatible percentage" and "Not compatible with new UK variant percentage".
- ^ In Austria, the listed B.1.1.7 variant shares represent the percentage of all positive COVID-19 tests being confirmed to be B.1.1.7 by genome sequencing. However, many N501Y positive RT-PCR tests were not variant determined further by genome sequencing, and likely would have returned a B.1.1.7 positive result for the vast majority of such tests if the genome sequencing analysis had been done. If all N501Y positive tests had been analysed further by genome sequencing, then the listed weekly B.1.1.7 shares could have been uptil: 9.6% for week 1 (2.4% higher), 23.8% for week 2 (6.4% higher), 19.6% for week 3 (4.9% higher), 28.6% for week 4 (6.1% higher), 37.2% for week 5 (8.5% higher), 45.4% for week 6 (21.4% higher), 58.1% for week 7 (23.0% higher), 65.3% for week 8 (4.0% higher), 68.0% for week 9 (6.8% higher) and 73.7% for week 10 (25.4% higher).
- In Sweden, a study comprising 11% of all SARS-CoV-2–positive PCR samples nationwide for week 4 found the B.1.1.7 share to be 10.8% (243/2244). Samples were, however, only collected from five southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro), which were not considered to be statistically representative for the demography and geography of Sweden as a whole. The national authorities plan to expand the weekly study to cover more regions for the following weeks in February 2021.
- In Sweden, a study comprising 16% of all SARS-CoV-2–positive PCR samples nationwide for week 5 found the B.1.1.7 share to be 15.1% (488/3224). Samples were, however, only collected from five southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro), which were not considered to be statistically representative for the demography and geography of Sweden as a whole. The national authorities also calculated a two-week average (week 5+6) for 19 out of 21 regions, and plan to expand the weekly study to cover more regions for the following weeks in February 2021.
- In Sweden, a study comprising 18% of all SARS-CoV-2–positive PCR samples nationwide for week 6 found the B.1.1.7 share to be 27.3% (1021/3742). Samples were, however, only collected from five southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro), which were not considered to be statistically representative for the demography and geography of Sweden as a whole. The national authorities also calculated a two-week average (week 5+6) for 19 out of 21 regions, and plan to expand the weekly study to cover more regions for the following weeks in February 2021.
- In Sweden, a study comprising 47.8% of all SARS-CoV-2–positive PCR samples nationwide for week 7, collected samples from 19 out of 21 regions (all except Gotland and Västerbotten), and found the B.1.1.7 share to be 30.4% (3316/10910) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).
- In Sweden, a study comprising 42.2% of all SARS-CoV-2–positive PCR samples nationwide for week 8, collected samples from 19 out of 21 regions (all except Gotland and Västerbotten), and found the B.1.1.7 share to be 41.5% (4643/11191) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).
- In Sweden, a study comprising 37.8% of all SARS-CoV-2–positive PCR samples nationwide for week 9, collected samples from 17 out of 21 regions (all except Gotland, Västerbotten, Norrbotten and Östergötland), and found the B.1.1.7 share to be 56.4% (5939/10528) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).
- In Sweden, a study comprising 43.1% of all SARS-CoV-2–positive PCR samples nationwide for week 10, collected samples from 18 out of 21 regions (all except Gotland, Västerbotten and Östergötland), and found the B.1.1.7 share to be 71.3% (8850/12417) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).
References
- ^ "B.1.1.7 report". cov-lineages.org. Retrieved 29 January 2021.
- "Tracking SARS-CoV-2 variants". www.who.int. Retrieved 17 August 2022.
- "Variants of concern". CDGN. Retrieved 17 August 2022.
- Kupferschmidt, Kai. "Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear". www.science.org. Science. Retrieved 24 February 2023.
- ^ Peacock, Sharon (22 December 2020). "Here's what we know about the new variant of coronavirus". The Guardian.
- Donnelly, Laura (26 January 2021). "UK to help sequence mutations of Covid around world to find dangerous new variants". The Telegraph. Archived from the original on 11 January 2022. Retrieved 28 January 2021.
- Rachel Schraer (22 December 2020). "Covid: New variant found 'due to hard work of UK scientists". BBC. Retrieved 30 January 2021.
- Sugden, Joanna (30 January 2021). "How the U.K. Became World Leader in Sequencing the Coronavirus Genome". The Wall Street Journal.
- ^ Investigation of novel SARS-CoV-2 variant Variant of Concern 202012/01: Technical briefing 5 (PDF) (Report). Public Health England. 2 February 2021. Retrieved 2 February 2021.
- ^ Public Health England (16 February 2021). "Variants: distribution of cases data". Gov.UK. Retrieved 17 February 2021.
- "Explained: Why WHO named Covid-19 variants first found in India as 'Kappa' and 'Delta' | India News - Times of India". The Times of India. 1 June 2021. Retrieved 2 June 2021.
- "WHO announces simple, easy-to-say labels for SARS-CoV-2 Variants of Interest and Concern". www.who.int. Retrieved 6 June 2021.
- For an extensive list of news sources using these terms, see UK Covid-19 variant, UK coronavirus variant and UK variant
- ^ Roberts, Michelle (2 February 2021). "UK variant has mutated again, scientists say". BBC News. Retrieved 2 February 2021.
- Boseley, Sarah (10 February 2021). "Mutated Kent Covid variant must be taken seriously, warns UK scientist". The Guardian. Retrieved 10 February 2021.
- ^ "'Has everyone in Kent gone to an illegal rave?': on the variant trail with the Covid detectives". the Guardian. 3 April 2021. Retrieved 23 September 2022.
- "PHE investigating a novel strain of COVID-19". Public Health England. 14 December 2020.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- "Variants: distribution of cases data". GOV.UK. 28 January 2021. At "Differences between a Variant of Concern and Variant Under Investigation". Retrieved 19 February 2021.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- ^ Rambaut, Andrew; Loman, Nick; Pybus, Oliver; Barclay, Wendy; Barrett, Jeff; Carabelli, Alesandro; et al. (19 December 2020). Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations (Report). Written on behalf of COVID-19 Genomics Consortium UK. Retrieved 20 December 2020.
- Callaway, Ewen (15 January 2021). "'A bloody mess': Confusion reigns over naming of new COVID variants". Nature News & Comment. 589 (7842): 339. Bibcode:2021Natur.589..339C. doi:10.1038/d41586-021-00097-w. PMID 33452513.
Emma Hodcroft, a molecular epidemiologist at the University of Bern, Switzerland, who is part of Nextstrain, the SARS-CoV-2 naming effort that called the 'UK variant' 20I/501Y.V1.
- Spike Variants: Alpha variant, aka B.1.1.7, 501Y.V1, 20I/501Y.V1, and UK COVID variant 24 June 2021, covdb.stanford.edu, accessed 1 July 2021
- ^ Wise, Jacqui (16 December 2020). "Covid-19: New coronavirus variant is identified in UK". BMJ. 371: m4857. doi:10.1136/bmj.m4857. PMID 33328153. S2CID 229291003.
- Chand et al., p. 5.
- "Headache and runny nose linked to Delta variant". BBC News. 14 June 2021.
- "Rapid evaluation confirms lateral flow devices effective in detecting new COVID-19 variant". Gov.UK. Retrieved 27 December 2020.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- Patel-Carstairs, Sunita (19 December 2020). "COVID-19: London and South East set for Tier 4 rules – as new COVID variant 'real cause for concern'". Sky News.
- "Covid vaccines 'still effective' against fast-spreading variant". Metro. 20 December 2020.
- "Vaccines effective against new virus strain – German health minister". Inquirer. AFP. 21 December 2020.
- ^ "NERVTAG meeting on SARS-CoV-2 variant under investigation: VUI-202012/01". New and Emerging Respiratory Virus Threats Advisory Group. 18 December 2020.
- ^ "COVID-19 (SARS-CoV-2): information about the new virus variant". Gov.uk. Public Health England. 20 December 2020. Retrieved 21 December 2020.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- Brown, Faye (9 February 2021). "Mutant Covid strain found in Bristol designated 'variant of concern'". Metro UK. Retrieved 10 February 2021.
- Mandavilli, Apoorva (5 March 2021). "In Oregon, Scientists Find a Virus Variant With a Worrying Mutation". The New York Times. ISSN 0362-4331. Archived from the original on 28 December 2021. Retrieved 6 March 2021.
- "B.1.1.7 Lineage with S:E484K Report". Outbreak. Retrieved 6 March 2021.
- Volz E, Mishra S, et al. (May 2021). "Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England". Nature. 593 (7858): 266–269. Bibcode:2021Natur.593..266V. doi:10.1038/s41586-021-03470-x. hdl:10044/1/87474. ISSN 1476-4687. PMID 33767447. S2CID 232365193.
- Davies NG, Abbott S, et al. (9 April 2021). "Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England". Science. 372 (6538): eabg3055. doi:10.1126/science.abg3055. PMC 8128288. PMID 33658326.
- Fort, Hugo (August 2021). "A very simple model to account for the rapid rise of the alpha variant of SARS-CoV-2 in several countries and the world". Virus Research. 304: 198531. doi:10.1016/j.virusres.2021.198531. PMC 8339456. PMID 34363849.
- Leung, Kathy; Shum, Marcus HH; Leung, Gabriel M; Lam, Tommy TY; Wu, Joseph T (7 January 2021). "Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020". Eurosurveillance. 26 (1). doi:10.2807/1560-7917.ES.2020.26.1.2002106. PMC 7791602. PMID 33413740.
- Kraemer MU, Hill V, et al. (22 July 2021). "Spatiotemporal invasion dynamics of SARS-CoV-2 lineage B.1.1.7 emergence". Science. 373 (6557): 889–895. Bibcode:2021Sci...373..889K. doi:10.1126/science.abj0113. hdl:10044/1/90624. ISSN 0036-8075. PMC 9269003. PMID 34301854. S2CID 236209853.
- Pinkstone, Joe (23 July 2021). "Scientists overestimated alpha variant's transmissibility". The Telegraph. ISSN 0307-1235. Archived from the original on 11 January 2022. Retrieved 10 August 2021.
- van Dissel, Jaap (24 February 2021). "COVID-19 2e Kamer-briefing, 24 febr 2021" [Technical 2nd chamber briefing on coronavirus developments, 24 February 2021] (PDF) (in Dutch). Retrieved 28 February 2021.
- ^ Ekspertrapport af den 21.februar 2021: Prognoser for smittetal og indlæggelser ved genåbningsscenarier d. 1. marts [Expert report of 21 February 2021: Forecasts for infection rates and hospital admissions for reopening scenarios 1 March] (PDF) (Report) (in Danish). Statens Serum Institut. 22 February 2021. Retrieved 22 February 2021.
- "Transmission of SARS-CoV-2 variants in Switzerland". Institute of Social and Preventive Medicine (ISPM), University of Bern. 5 March 2021. Retrieved 5 March 2021.
- ^ Volz E, Mishra S, et al. (4 January 2021). "Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data". medRxiv 10.1101/2020.12.30.20249034.
- "New evidence on VUI-202012/01 and review of the public health risk assessment". Retrieved 4 January 2021.
- "COG-UK Showcase Event". YouTube. Retrieved 25 December 2020.
- Grabowski, Frederic; Preibisch, Grzegorz; Giziński, Stanisław; Kochańczyk, Marek; Lipniacki, Tomasz (1 March 2021). "SARS-CoV-2 Variant of Concern 202012/01 Has about Twofold Replicative Advantage and Acquires Concerning Mutations". Viruses. 13 (3): 392. doi:10.3390/v13030392. PMC 8000749. PMID 33804556.
- "UniProtKB - P0DTC2 (SPIKE_SARS2)". UniProt. Retrieved 4 February 2021.
- ^ National Public Health Emergency Team, Department of Health (25 February 2021). "NPHET COVID-19 Update – 25 February 2021" (PDF). New (B.1.1.7) variant - S gene target failure (p 31). Retrieved 4 March 2021.
{{cite web}}: CS1 maint: numeric names: authors list (link) - Pylas, Pan (2 January 2021). "U.K. breaks record with 57,725 cases, is urged to keep schools closed". Coronavirus. Retrieved 3 January 2021.
- Mahase, Elisabeth (23 December 2020). "Covid-19: What have we learnt about the new variant in the UK?". BMJ. 371: m4944. doi:10.1136/bmj.m4944. PMID 33361120. S2CID 229366003.
- Kirby, Tony (February 2021). "New variant of SARS-CoV-2 in UK causes surge of COVID-19". The Lancet Respiratory Medicine. 9 (2): e20–e21. doi:10.1016/S2213-2600(21)00005-9. PMC 7784534. PMID 33417829.
- ^ COG-UK update on SARS-CoV-2 Spike mutations of special interest: Report 1 (PDF) (Report). COVID-19 Genomics UK Consortium (COG-UK). 20 December 2020. p. 7. Archived from the original (PDF) on 22 December 2020.
- COG-UK (20 December 2020), p. 4.
- Yi, C.; Sun, X.; Ye, J.; et al. (2020). "Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies". Cellular & Molecular Immunology. 17 (6): 621–630. doi:10.1038/s41423-020-0458-z. PMC 7227451. PMID 32415260.
- ^ COG-UK (20 December 2020), p. 1.
- ^ Chand et al., p. 6.
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; et al. (2021). "SARS-CoV-2 evolution during treatment of chronic infection". Nature (Accelerated article preview). 592 (7853): 277–282. doi:10.1038/s41586-021-03291-y. PMC 7610568. PMID 33545711. S2CID 232113127.
- Shahhosseini, Nariman; Babuadze, George (Giorgi); Wong, Gary; Kobinger, Gary P. (May 2021). "Mutation Signatures and In Silico Docking of Novel SARS-CoV-2 Variants of Concern". Microorganisms. 9 (5): 926. doi:10.3390/microorganisms9050926. PMC 8146828. PMID 33925854.
- Barton, Michael I; MacGowan, Stuart A; Kutuzov, Mikhail A; Dushek, Omer; Barton, Geoffrey John; van der Merwe, P Anton (26 August 2021). Fouchier, Ron AM; Van der Meer, Jos W; Fouchier, Ron AM (eds.). "Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics". eLife. 10: e70658. doi:10.7554/eLife.70658. ISSN 2050-084X. PMC 8480977. PMID 34435953.
- Challen R, Brooks-Pollock E, et al. (9 March 2021). "Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study". BMJ. 372: n579. doi:10.1136/bmj.n579. PMC 7941603. PMID 33687922.
- ^ Davies NG, Jarvis CI, et al. (13 May 2021). "Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7". Nature. 593 (7858): 270–274. Bibcode:2021Natur.593..270D. doi:10.1038/s41586-021-03426-1. ISSN 0028-0836. PMC 9170116. PMID 33723411. S2CID 232245304.
- Frampton D, Rampling T, et al. (April 2021). "Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study". The Lancet Infectious Diseases. 21 (9): 1246–1256. doi:10.1016/S1473-3099(21)00170-5. PMC 8041359. PMID 33857406.
- Graham MS, Sudre CH, et al. (May 2021). "Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study". The Lancet Public Health. 6 (5): e335–e345. doi:10.1016/S2468-2667(21)00055-4. PMC 8041365. PMID 33857453.
- ^ "Covid: WHO in 'close contact' with UK over new virus variant". BBC News. 20 December 2020.
- Berger, Miriam (20 December 2020). "Countries across Europe halt flights from Britain over concerns about coronavirus mutation". Washington Post. ISSN 0190-8286. Archived from the original on 20 December 2020. Retrieved 20 December 2020.
- "New UK variant 'may be more deadly'". BBC News. 22 January 2021.
- Peter Horby; Catherine Huntley; Nick Davies; John Edmunds; Neil Ferguson; Graham Medley; Andrew Hayward; Muge Cevik; Calum Semple (11 February 2021). "NERVTAG paper on COVID-19 variant of concern B.1.1.7: NERVTAG update note on B.1.1.7 severity (2021-02-11)" (PDF). www.gov.uk.
- Peston, Robert (22 January 2021). "New Covid-19 strain may be more lethal, Robert Peston learns". ITV News. Retrieved 24 January 2021.
- Challen, Robert; Brooks-Pollock, Ellen; Read, Jonathan M; Dyson, Louise; Tsaneva-Atanasova, Krasimira; Danon, Leon (9 March 2021). "Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study". BMJ. 372: n579. doi:10.1136/bmj.n579. PMC 7941603. PMID 33687922.
- Statens Serum Institut (24 February 2021). "B.1.1.7 kan føre til flere indlæggelser" [B.1.1.7 might lead to more hospital admissions] (in Danish). Retrieved 25 February 2021.
- ^ Zinzula, Luca (2020). "Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2". Biochemical and Biophysical Research Communications. 538: 116–24. doi:10.1016/j.bbrc.2020.10.045. PMC 7577707. PMID 33685621.
- Kupferschmidt, Kai (20 December 2020). "Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear". Science. doi:10.1126/science.abg2626. S2CID 234564870.
See also COG-UK (20 December 2020), p. 4: "69–70del has been identified in variants associated with immune escape in immunocompromised patients...." - "Genomic overview of SARS-CoV-2 in Denmark". 30 January 2021. Retrieved 1 February 2021.
- ^ National Institute for Public Health and the Environment (RIVM) - Ministry of Health, Welfare and Sport (23 March 2021). "Variants of the coronavirus SARS-CoV-2". www.rivm.nl. Retrieved 23 March 2021.
- ^ Borges, Vítor; et al. (19 January 2021). "Tracking SARS-CoV-2 VOC 202012/01 (lineage B.1.1.7) dissemination in Portugal: insights from nationwide RT-PCR Spike gene drop out data". Update information including data from weeks 3-10, 2021: Posted after the article on 16 March 2021. Footnote of the previous updates explain that 91.8% of total SGTF+SGTL (~90% of SGTF and 100% of SGTL) was confirmed by genome sequence to be the B.1.1.7 variant. The B.1.1.7 shares were calculated as 91.8% of the total SGTF+SGTL share for each week. Retrieved 16 March 2021.
- ^ Swiss National COVID-19 Science Task Force. "Genomic Characterisation". Retrieved 15 March 2021.
{{cite web}}: CS1 maint: numeric names: authors list (link) - ^ "SARS-CoV-2 Variants of Concern in Switzerland". IBZ Shiny. January 2021. Archived from the original on 17 February 2021. Retrieved 18 February 2021.
- Data sources:
- Denmark:
- The Netherlands:
- Portugal:
- Switzerland:
- The United Kingdom:
- Ireland:
- ^ "Coronavirus (COVID-19) Infection Survey, UK: 29 January 2021: 10. Positive tests that are compatible with the new UK variant". www.ons.gov.uk. 29 January 2021. Retrieved 1 February 2021.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- Walker, A. Sarah; Vihta, Karina-Doris; Gethings, Owen; Pritchard, Emma; Jones, Joel; House, Thomas; Bell, Iain; Bell, John I; Newton, John N; Farrar, Jeremy; Diamond, Ian; Studley, Ruth; Rourke, Emma; Hay, Jodie; Hopkins, Susan; Crook, Derrick; Peto, Tim; Matthews, Philippa C.; Eyre, David W.; Stoesser, Nicole; Pouwels, Koen B. (15 January 2021). "Increased infections, but not viral burden, with a new SARS-CoV-2 variant". medRxiv 10.1101/2021.01.13.21249721.
- Data sources:
- UK national (2021-01-21):
- South-East England (2020-12-31):
- UK countries (2021-01-29):
- England regions (2021-01-15):
- "Risk related to spread of new SARSCoV-2 variants of concern in the EU/EEA" (PDF). www.ecdc.europa.eu. 29 December 2020. Retrieved 1 February 2021.
- ^ Statens Serum Institut (4 February 2021). "Delta-PCR-testen" [The delta-PCR test] (in Danish). Retrieved 11 February 2021.
- Antonin Bal; Gregory Destras; Alexandre Gaymard; Karl Stefic; Julien Marlet; et al. (COVID-Diagnosis HCL Study Group) (21 January 2021). "Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69–V70; France; August to December 2020". Eurosurveillance. 26 (3). doi:10.2807/1560-7917.ES.2021.26.3.2100008. PMC 7848679. PMID 33478625.
- ^ Santé Publique France (11 March 2021). "COVID-19: Point épidémiologi que hebdomadaire du 11 mars 2021" [COVID-19: Weekly epidemiological update of 11 March 2021] (in French). Retrieved 12 March 2021.
- ^ "Statistik om SARS-CoV-2 virusvarianter av särskild betydelse" [Statistics on SARS-CoV-2 Variants Of Concern] (in Swedish). Folkhälsomyndigheten. 18 March 2021. Retrieved 18 March 2021.
- ^ "COVID-19 weekly epidemiological update, 23 March 2021" (PDF). WHO. 23 March 2021. Retrieved 23 March 2021.
- Younes M, Hamze K, et al. (26 January 2021). "Emergence and fast spread of B.1.1.7 lineage in Lebanon". medRxiv 10.1101/2021.01.25.21249974.
- Younes M, Hamze K, et al. (17 March 2021). "B.1.1.7 became the dominant variant in Lebanon". medRxiv 10.1101/2021.03.17.21253782.
- Higgins-Dunn, N. (19 December 2020). "The U.K. has identified a new Covid-19 strain that spreads more quickly. Here's what they know". MSNBC.
- Team, The Lockdown Files (9 March 2023). "Covid variant that ruined Christmas hidden by scientists from ministers for months". The Telegraph. ISSN 0307-1235. Retrieved 13 March 2023.
- "COVID-19 Genomics UK (COG-UK) Consortium - Wellcome Sanger Institute". www.sanger.ac.uk. Retrieved 23 December 2020.
- Gallagher, James (20 December 2020). "New coronavirus variant: What do we know?". BBC News. Retrieved 21 December 2020.
- Ross, T.; Spence, E. (19 December 2020). "London Begins Emergency Lockdown as U.K. Fights New Virus Strain". Bloomberg News.
- "Variants – distribution of cases data: data up to 25 January 2021". Gov.uk. Public Health England. 26 January 2021. Retrieved 27 January 2021.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- ^ Public Health England (1 February 2021). "Investigation of novel SARS-CoV-2 variant: 202012/01. Technical briefing 5 data". Data report -"Figure 1", shows the weekly percentage of all genome sequenced VOC202012/01 by adding the SGTF+Kent share with the Not SGTF+Kent share. The report use the word "Kent" as the abbreviated nickname for VOC202012/01. Retrieved 5 February 2021.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- ^ "(09.03.2021 Г.) ЕПИДЕМИЯ ОТ ОСТЪР РЕСПИРАТОРЕН СИНДРОМ, СВЪРЗАНА С НОВ КОРОНАВИРУС SARS-COV-2, УХАН, КИТАЙ: АКТУАЛНА ИНФОРМАЦИЯ" [(09.03.2021) Acute Respiratory Syndrome Epidemic Associated With New Coronavirus SARS-COV-2, Wuhan, China: Actual Information] (in Bulgarian). 3 March 2021. Retrieved 10 March 2021.
- Kovacova, V.; et al. (5 February 2021). "A novel, room temperature-stable, multiplexed RT-qPCR assay to distinguish lineage B.1.1.7 from the remaining SARS-CoV-2 lineages". virological.org. Retrieved 21 February 2021.
- ^ Ján Mikas, hlavný hygienik Slovenskej Republiky (9 March 2021). "Britský variant potvrdený na Slovensku v 90% vyšetrených vzoriek" [British variant confirmed in Slovakia in 90% of examined samples] (in Slovak). Public Health Office of the Slovak Republic. Archived from the original on 13 May 2021. Retrieved 17 March 2021.
- "Aktuálna epidemiologická situácia na Slovensku (prezentácia, 8. január 2021)" [Current epidemiological situation in Slovakia (presentation, January 8, 2021)] (PDF) (in Slovak). Public Health Office of the Slovak Republic. 9 January 2021. Archived from the original (PDF) on 14 July 2021. Retrieved 21 February 2021.
- "Press Releases: Four Cases of COVID-19 Variant found in the United Kingdom Have Been Discovered in Israel". Ministry of Health. 23 December 2020. Retrieved 26 February 2021.
- ^ Adrian Pilot (5 January 2021). "המוטציה הבריטית נמצאת ב-10% עד 20% ממאומתי הקורונה" [The British mutation is 10% to 20% of the corona nationwide] (in Hebrew). Calcalist. Retrieved 26 February 2021.
- ^ Toi Staff (2 February 2021). "We will have to live with COVID-19 for a long time, says Israeli expert". Times of Israel. Retrieved 26 February 2021.
- ^ Ynet (16 February 2021). "Senior health official: 90% of COVID-19 cases caused by UK variant". Retrieved 25 February 2021.
- ^ Laboratoire National de Santé (LNS) (18 March 2021). "Respiratory Viruses Surveillance – REVILUX: Respiratory Viruses Sentinelle Newsletter, REVILUX bulletin hebdomadaire" [Respiratory Viruses Surveillance – REVILUX: Respiratory Viruses Sentinelle Newsletter, REVILUX weekly update]. Archived from the original on 17 December 2021. Retrieved 18 March 2021.
- ^ "Status for udvikling af SARS-CoV-2 Variants of Concern (VOC) i Danmark" [Status of development of SARS-CoV-2 Variants of Concern (VOC) in Denmark] (in Danish). Statens Serum Institut. 20 March 2021. Retrieved 20 March 2021.
- Notat om prognoser for smittetal og indlæggelser ved scenarier for genåbning af 0.-4. klasse i grundskolen [Memorandum on forecasts for infection rates and hospital admissions in scenarios for reopening of 0th–4th class in primary school] (PDF) (Report) (in Danish). Statens Serum Institut. 31 January 2021. Retrieved 2 February 2021.
- "Status for udvikling af SARS-CoV-2 Variants of Concern (VOC) i Danmark" [Status of development of SARS-CoV-2 Variants of Concern (VOC) in Denmark] (in Danish). Statens Serum Institut. 12 March 2021. Retrieved 20 March 2021.
- "Status for udvikling af SARS-CoV-2 Variants of Concern (VOC) i Danmark" [Status of development of SARS-CoV-2 Variants of Concern (VOC) in Denmark] (in Danish). Statens Serum Institut. 19 March 2021. Retrieved 20 March 2021.
- Ministerie van Volksgezondheid, Welzijn en Sport (23 February 2021). "Advies VWS na OMT 101" [Advice from VWS after 101th OMT on COVID-19] (PDF). overheid.nl (in Dutch). Retrieved 13 April 2022.
- ^ Folkehelseinstituttet (3 March 2021). "COVID-19 Ukerapport – uke 8, onsdag 3. mars 2021" [COVID-19 weekly report for week 8 (wednesday 3 March 2021)] (PDF) (in Norwegian). Archived from the original (PDF) on 19 November 2021. Retrieved 4 March 2021.
- ^ Folkehelseinstituttet (17 March 2021). "COVID-19 Ukerapport – uke 10, onsdag 17. mars 2021" [COVID-19 weekly report for week 10 (wednesday 17 March 2021)] (PDF) (in Norwegian). Archived from the original (PDF) on 17 March 2021. Retrieved 17 March 2021.
- Folkehelseinstituttet (26 February 2021). "Statistikk over meldte tilfeller av den engelske og den sør-afrikanske varianten av koronavirus" [Statistics on reported cases of the English and South African variants of coronavirus] (in Norwegian). Retrieved 28 February 2021.
- Folkehelseinstituttet (27 February 2021). "Engelsk virusvariant dominerer i Oslo og Viken" [English virus variant dominates in Oslo and Viken] (in Norwegian). Archived from the original on 17 April 2021. Retrieved 28 February 2021.
- Instituto Nacional de Saúde (INSA), Doutor Ricardo Jorge (5 February 2021). "Diversidade genética do novo coronavírus SARS-CoV-2 (COVID-19) em Portugal" [Genetic diversity of the new SARS-CoV-2 coronavirus (COVID-19) in Portugal] (PDF) (in Portuguese). Retrieved 7 February 2021.
- Instituto Nacional de Saúde (INSA), Doutor Ricardo Jorge (3 March 2021). "Diversidade genética do novo coronavírus SARS-CoV-2 (COVID-19) em Portugal (3 de Março de 2021)" [Genetic diversity of the new SARS-CoV-2 coronavirus (COVID-19) in Portugal (3 March 2021)] (PDF) (in Portuguese). Retrieved 4 March 2021.
- ^ Istituto Superiore di Sanità (15 February 2021). "Prevalenza della variante VOC 202012/01, lineage B.1.1.7 in Italia (Studio di prevalenza 4-5 febbraio 2021)" [Prevalence of the VOC variant 202012/01, lineage B.1.1.7 in Italy (Prevalence study February 4–5, 2021)] (in Italian). Retrieved 17 February 2021.
- ^ Istituto Superiore di Sanità (2 March 2021). "CS N° 14/2021 - In Italia il 54% delle infezioni dovute a 'variante inglese', il 4,3% a quella 'brasiliana' e lo 0,4% a quella 'sudafricana'" [CS N° 14/2021 - In Italy 54% of infections due to the 'English variant', 4.3% to the 'Brazilian' one and 0.4% to the 'South African' one] (in Italian). Retrieved 5 March 2021.
- "Wissenschaftliches update 09 februar 2021" [Scientific update of 9 February 2021] (in German). Swiss National COVID-19 Task Force. 9 February 2021. Retrieved 17 February 2021.
- ^ Sciensano (19 March 2021). "Covid-19 Wekelijks Epidemiologisch Bulletin (19 Maart 2021)" [Covid-19 Weekly Epidemiological Bulletin (19 March 2021)] (PDF). Moleculaire surveillance van SARS-CoV-2 (p.20-22) (in Dutch). Retrieved 19 March 2021.
- ^ Sciensano (19 February 2021). "Covid-19 Wekelijks Epidemiologisch Bulletin (19 Februari 2021)" [Covid-19 Weekly Epidemiological Bulletin (February 19, 2021)] (PDF). Moleculaire surveillance van SARS-CoV-2 (p.21-24) (in Dutch). Retrieved 25 February 2021.
- "Estimated date of dominance of VOC-202012/01 strain in France and projected scenarios" (PDF). 16 January 2021.
- ^ Santé Publique France (28 January 2021). "COVID-19: Point épidémiologi que hebdomadaire du 28 janvier 2021" [COVID-19: Weekly epidemiological update of 28 January 2021] (in French). Retrieved 1 February 2021.
- ^ Santé Publique France (4 March 2021). "COVID-19: Point épidémiologi que hebdomadaire du 4 mars 2021" [COVID-19: Weekly epidemiological update of 4 March 2021] (in French). Retrieved 5 March 2021.
- ^ Santé Publique France (18 March 2021). "COVID-19: Point épidémiologi que hebdomadaire du 18 mars 2021" [COVID-19: Weekly epidemiological update of 18 March 2021] (in French). Retrieved 21 March 2021.
- ^ "SARS-CoV-2-Varianten in Österreich" (in German). Österreichische Agentur für Gesundheit und Ernährungssicherheit GmbH (AGES). 19 March 2021. Retrieved 19 March 2021.
- ^ "Aktualisierter Bericht zu Virusvarianten von SARS-CoV-2 in Deutschland, insbesondere zur Variant of Concern (VOC) B.1.1.7: Stand: 17. März 2021" [Updated report on virus variants of SARS-CoV-2 in Germany, in particular on Variant of Concern (VOC) B.1.1.7: As of 17 March 2021] (PDF). www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/DESH/Berichte-VOC-tab.html (in German). Robert Koch-Institut (Bundesinstitut im Geschäftsbereich des Bundesministeriums für Gesundheit). 17 March 2021. Retrieved 20 March 2021.
- ^ "Three cases of new COVID variant detected in Malta". Times of Malta. 30 December 2020.
- Laura Calleja (24 February 2021). "COVID-19: Bars and band clubs to remain shut in March, Chris Fearne says". Malta Today. Retrieved 13 March 2021.
- "Malta to get new swab tests which can identify Covid-19 variants". Malta Independent. 4 March 2021. Retrieved 13 March 2021.
- ^ Claire Farrugia (10 March 2021). "UK virus variant detected in 60 per cent of new COVID cases". Times of Malta. Retrieved 10 March 2021.
- Folkhälsomyndigheten (4 February 2021). "Scenarier för fortsatt spridning – interimsrapport" [Scenarios for continued dissemination - interim report] (in Swedish). Retrieved 8 February 2021.
- ^ "Publicerad statistik för SARS-CoV-2 virusvarianter av särskild betydelse under vecka 8 2021" [Statistics on SARS-CoV-2 Variants Of Concern for week 8 2021] (in Swedish). Folkhälsomyndigheten. 15 March 2021. Archived from the original on 11 October 2021. Retrieved 15 March 2021.
- ^ "COVID-19 Switzerland Information on the current situation, as of 18 March 2021: Key figures, Liechtenstein". Download data: Data as .csv file. Federal Office of Public Health. 18 March 2021. Retrieved 18 March 2021.
- ^ "Spanish Health Ministry estimates British variant accounts for up to 10% of positives in Spain". murciatoday.com. 30 January 2021. Retrieved 3 February 2021.
- Alvaro Soto (18 February 2021). "La variante británica representa ya el 20% de los casos en España" [The British variant already represents 20% of cases in Spain]. msn.com (in Spanish). Retrieved 22 February 2021.
- ^ "Autoridades sanitarias afirmaron que la cepa británica circula "ampliamente" en España" [Health authorities affirmed that the British strain circulates "widely" in Spain] (in Spanish). Télam. 18 February 2021. Retrieved 17 March 2021.
- ^ Manuel Vilaseró (22 February 2021). "Detectadas en España las nuevas variantes nigeriana y de Río de Janeiro del coronavirus" [The new Nigerian and Rio de Janeiro variants of the coronavirus detected in Spain] (in Spanish). Retrieved 1 March 2021.
- ^ Miguel Hidalgo Pérez (3 March 2021). "Darias: "La variante británica ya es la dominante en España"" [Darias: "The British variant is already the dominant one in Spain"] (in Spanish). Retrieved 17 March 2021.
- ^ "Actualización de la situación epidemiológica de las variantes de importancia en salud pública en España (18 de marzo de 2021)" [Update of the epidemiological situation of Variants of Importance in Public Health in Spain (18 March 2021)] (PDF) (in Spanish). Ministerio de Sanidad, Consumo y Bienestar Social. 18 March 2021. Retrieved 18 March 2021.
- Adrian Dabek (11 February 2021). "Brytyjski wariant koronawirusa - jaka jest skala jego obecności w Polsce?" [British variant of the coronavirus - what is the scale of its presence in Poland?] (in Polish). Medonet. Retrieved 17 February 2021.
- European Centre for Disease Prevention and Control (15 February 2021). "Rapid Risk Assessment: SARS-CoV-2 increased circulation of variants of concern and vaccine rollout in the EU/EEA, 14th update, 15 February 2021" (PDF). www.ecdc.europa.eu. Retrieved 18 February 2021.
- "Od 20 marca w całej Polsce obowiązują rozszerzone zasady bezpieczeństwa" [From March 20, extended safety rules apply throughout Poland]. gov.pl (in Polish). Government of Poland. 17 March 2021. Retrieved 17 March 2021.
- ^ Sylwia Dabkowska-Pozyczka; Edyta Ros; Wiktoria Nicalek; Karol Kostrzewa; Aleksandra Rebelinska (17 March 2021). "Nowe obostrzenia w calym kraju od 20 marca do 9 kwietnia" [New restrictions nationwide from March 20 to April 9]. pap.pl (in Polish). Polska Agencja Prasowa. Retrieved 17 March 2021.
- Piotr Kallalas (2 March 2021). "Brytyjski wariant koronawirusa może dominować na Pomorzu" [The British variant of the coronavirus may be dominant in Pomerania]. trojmiasto.pl (in Polish). Retrieved 17 March 2021.
- "Koronawirus. Brytyjski wariant w Wielkopolsce. Najnowsze dane" [Coronavirus. British variant in Greater Poland. Latest data]. Onet.pl (in Polish). 17 March 2021. Retrieved 17 March 2021.
- Finish Institute for Health and Welfare, TLH (17 February 2021). "Uppföljning av hybridstrategin för covid-19-epidemin: Separat översikt, varierande tema (Uppföljning av muterade coronavirus 17.2.2021)" [Follow-up of the hybrid strategy for the COVID-19 epidemic: Separate overview, varying theme (Follow-up of mutated coronavirus 17.2.2021)] (in Swedish). Archived from the original on 14 July 2021. Retrieved 2 March 2021.
- "UK Covid variant spreading in Finland". Yle. 14 February 2021. Retrieved 2 March 2021.
- Jelena Ćirić (4 March 2021). "COVID-19 in Iceland: South African Variant Detected at Border". Iceland Review. Retrieved 13 March 2021.
- Washington NL, Gangvarapu K, et al. (7 February 2021). "Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States". medRxiv 10.1101/2021.02.06.21251159v1.
- CDC (28 March 2020). "COVID Data Tracker". Centers for Disease Control and Prevention. Retrieved 30 April 2021.
- Galloway SE, Paul P, et al. (22 January 2021). "Emergence of SARS-CoV-2 B.1.1.7 Lineage — United States, December 29, 2020–January 12, 2021". Morbidity and Mortality Weekly Report. 70 (3): 95–99. doi:10.15585/mmwr.mm7003e2. PMC 7821772. PMID 33476315.
- "More Covid Mysteries". New York Times. 30 July 2021.
- Aziz, Saba (27 December 2020). "Canada reports first cases of U.K. coronavirus variant. Here's what you need to know". Global News. Retrieved 14 February 2021.
- Thompson, Nicole (13 February 2021). "Contagious variant of COVID-19 now identified in all 10 provinces". The StarPhoenix. The Canadian Press. Retrieved 14 February 2021.
- ^ "Why Alberta has identified more variant cases but they're believed to be more prevalent in Ontario | CBC News". CBC. Retrieved 1 April 2021.
- "SARS-CoV-2 Variants of Concern" (PDF). Alberta Health Services.
- "Variants of Concern (VOC) in Canada". Government of Canada. 19 April 2020. Retrieved 24 March 2021.
- "SARS-CoV-2 Variants of Concern: Results of Point Prevalence Study" (PDF). Public Health Ontario. 12 February 2021. Retrieved 13 March 2021.
- Barbara Yaffe (16 February 2021). "Coronavirus: Ontario health official says approximately 7% of screened cases are due to variants in province". Global News. Retrieved 13 March 2021.
- "Ontario Dashboard: Unmasking the New Variants of Concern". Ontario COVID-19 Science Advisory Table. 16 March 2021. Archived from the original on 17 March 2021. Retrieved 16 March 2021.
- Kalina Laframboise (17 February 2021). "More contagious COVID-19 variant could become dominant strain in Montreal by March: INSPQ". Global News. Retrieved 13 March 2021.
- Public Health England (11 March 2021). "Investigation of SARS-CoV-2 variants of concern in England: Technical briefing 7" (PDF). Weekly number and proportion of England Pillar 2 COVID-19 cases with SGTF (Figure 10). Retrieved 12 March 2021.
- Public Health England (13 February 2021). "Investigation of SARS-CoV-2 variants of concern in England: Technical briefing 6" (PDF). Percentage of Pillar 2 Δ69-70 sequences that are VOC 202012/01 (Table 7), with a value of 100% since week 4 in 2021. Retrieved 14 February 2021.
- ^ Public Health England (19 March 2021). "Dataset: Coronavirus (COVID-19) Infection Survey". Table 6A: Percentage and Cycle threshold (Ct) values of COVID-19 cases, UK. Table 6C: New UK variant compatible, not compatible with new UK variant and virus too low for variant to be identifiable positives modelled daily estimates, UK. Retrieved 19 March 2021.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- ^ Public Health England (19 March 2021). "Coronavirus (COVID-19) Infection Survey, UK: 19 March 2021". Retrieved 19 March 2021.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- Ido Efreti (12 January 2021). "Coronavirus Live: Israel Sees Drop in Infections 14 Days After First Vaccine Dose (4:50 P.M. Mutation constitutes up to 20% of Israel's recent COVID-19 infections)". Haaretz. Retrieved 26 February 2021.
- "Senior health official: UK virus variant putting pregnant women at greater risk". www.timesofisrael.com. 21 January 2021. Retrieved 23 January 2021.
- Stuart Winer (25 January 2021). "Health officials: UK variant running wild, hitting children at a worrying rate". Times of Israel. Retrieved 25 February 2021.
- Maayan Lubell; Ari Rabinovitch (9 February 2021). "Vaccine vs variant: Promising data in Israel's race to defeat pandemic". Reuters. Retrieved 26 February 2021.
- Folkehelseinstituttet (24 February 2021). "COVID-19 Ukerapport – uke 7, onsdag 24. februar 2021" [COVID-19 weekly report for week 7 (wednesday 24 February 2021)] (PDF) (in Norwegian). Archived from the original (PDF) on 6 May 2021. Retrieved 28 February 2021.
- Santé Publique France (18 February 2021). "COVID-19: Point épidémiologi que hebdomadaire du 18 février 2021" [COVID-19: Weekly epidemiological update of February 18, 2021] (in French). Retrieved 20 February 2021.
- ^ Santé Publique France (25 February 2021). "COVID-19: Point épidémiologi que hebdomadaire du 25 février 2021" [COVID-19: Weekly epidemiological update of February 25, 2021] (in French). Retrieved 26 February 2021.
- "Stickprov tyder på ökad spridning av den brittiska virusvarianten i Sverige" [Random samples indicate an increased spread of the British virus variant in Sweden] (in Swedish). Folkhälsomyndigheten. 2 February 2021. Archived from the original on 12 May 2021. Retrieved 2 February 2021.
- ^ Helix. "The Helix® COVID-19 Surveillance Dashboard". Trends in S Gene Target Failure (SGTF), interactive data map: B.1.1.7 weekly share calculated as the average of the daily B.1.1.7 shares of the week. Daily B.1.1.7 shares are found by multiplying "Daily Percent SGTF of Positive Samples" and "B.1.1.7 as % of sequenced SGTF positives". Retrieved 17 March 2021.
- ^ "Tracking of variants: VUI202012/01 GR/501Y.V1 (B.1.1.7)". GISAID. Retrieved 19 February 2021.
- ^ Henley, Jon; Jones, Sam; Giuffrida, Angela; Holmes, Oliver (20 December 2020). "EU to hold crisis talks as countries block travel from UK over new Covid strain". The Guardian.
- Hope, Alan (20 December 2020). "Netherlands bans flights from UK over new Covid mutation". The Brussels Times.
- "Coronavirus, in Italia un soggetto positivo alla variante inglese" [Coronavirus, one person tests positive in Italy for the English variant]. la Repubblica (in Italian). 20 December 2020.
- Kelland, Kate (21 December 2020). "Explainer - The new coronavirus variant in Britain: How worrying is it?". Reuters.
- Squires, Nick; Orange, Richard (21 December 2020). "New coronavirus strain detected around the globe, from Gibraltar to Australia". The Telegraph. Archived from the original on 11 January 2022.
- "Singapore confirms first case of new Covid-19 strain from UK, a 17-year-old student who recently returned from Britain". The Straits Times. 23 December 2020. Retrieved 24 December 2020.
- "Israel confirms four cases of new Covid variant". The Guardian. 23 December 2020.
- Moriarty, Gerry (23 December 2020). "First case of UK variant strain of Covid-19 confirmed in Northern Ireland". The Irish Times. Retrieved 24 December 2020.
- Burger, Ludwig (24 December 2020). "Germany reports first case of coronavirus variant spreading in Britain". Reuters.
- "COVID-19: Nouvelle variante du coronavirus découverte dans deux échantillons en Suisse" [COVID-19: New variant of the coronavirus discovered in two samples in Switzerland] (in French). Federal Office of Public Health (Switzerland). 24 December 2020.
- Moloney, Eoghan (25 December 2020). "New UK variant of Covid-19 confirmed in Ireland while 1,025 new cases and two further deaths confirmed". Irish Independent. Retrieved 25 December 2020.
- Graham-Harrison, Emma (25 December 2020). "Japan reports five cases of coronavirus variant found in UK". The Guardian.
- "Ontario identifies first cases of COVID-19 U.K. variant in the province". Canadian Broadcasting Corporation (CBC News). 26 December 2020.
- "Coronavirus: More cases of new Covid variant found in Europe". BBC. 26 December 2020. Retrieved 26 December 2020.
- "France, Lebanon confirm first cases of new coronavirus variant". Aljazeera. 26 December 2020. Retrieved 26 December 2020.
- "Fall av den brittiska virusvarianten upptäckt i Sverige" [Cases of the British virus variant discovered in Sweden] (in Swedish). SVT. 26 December 2020.
- "Norway, Portugal confirm first cases of coronavirus variant in travellers from the UK". Newshub. 27 December 2020. Archived from the original on 30 December 2020. Retrieved 28 December 2020.
- "Jordan detects two coronavirus variant cases: minister". Gulf News. 27 December 2020. Retrieved 28 December 2020.
- "New UK variant Covid strain detected in Finland". Yle News. 28 December 2020.
- "South Korea reports cases of COVID variant - and says they came from UK". Sky News. 28 December 2020.
- "Chile records first case of British variant of coronavirus - health ministry". Reuters. 29 December 2020.
- "Coronavirus: India confirms six cases of new Covid variant". BBC News. 29 December 2020.
- "First confirmed case of new Covid-19 strain detected in Pakistan". Hindustan Times. 29 December 2020.
- "New Covid strain in UAE: All we know so far". Khaleej Times. 30 December 2020.
- "Taiwan reports its first case of mutant Covid-19 strain found in Britain". South China Morning Post. 30 December 2020.
- "China confirms first case of UK coronavirus variant". France24. 31 December 2020.
- "Brazil detects two cases of new coronavirus variant found in UK". Reuters. 31 December 2020.
- Knudsen, T.H. (20 December 2020). "Dansk Oxford-professor: Danmark skal gøre alt for, at ny virusvariant ikke spreder sig" [Danish Oxford professor: Denmark must do everything to ensure that new virus variant does not spread] (in Danish). DR.
See also: "Global sequencing coverage". covidcg.org. Retrieved 23 December 2020. - Mandavilli, Apoorva; Landler, Mark; Castle, Stephen (20 December 2020). "Scientists urge calm about coronavirus mutations, which are not unexpected". New York Times.
- Casiano, Louis (29 December 2020). "Colorado health officials confirm new COVID-19 variant in the state". Fox News. Retrieved 29 December 2020.
- Nedelman, Michael (6 January 2021). "CDC has found more than 50 US cases of coronavirus variant first identified in UK". CNN. Retrieved 7 January 2021.
- Reimann, Nicholas (8 January 2021). "'Close To A Worst-Case Scenario'—Former CDC Director Issues 'Horrifying' Outlook For New Covid Strain". Forbes. Retrieved 15 January 2021.
- "Son dakika haberi... Bakan Koca açıkladı, 15 kişide mutasyonlu virüs! İngiltere'den girişler tamamen durduruldu" [Breaking news... Minister Koca announced, mutated virus in 15 people! Entries from the UK are completely suspended] (Video). CNN Türk. Retrieved 4 January 2021.
- "SSI: Meget smitsom coronamutation fra England spreder sig i Danmark" [SSI: Very infectious corona mutation [sic] from England spreads in Denmark]. DR (in Danish). 2 January 2021. Retrieved 3 January 2021.
- "Udvikling i smitte med engelsk virusvariant af SARS-COV-2 (cluster B.1.1.7)" [Development in infection with English virus variant of SARS-COV-2 (cluster B.1.1.7)] (PDF) (in Danish). Statens Serum Institut. 1 January 2021. Retrieved 2 January 2021.
- "Status for udvikling af B.1.1.7 i Danmark d. 17. januar 2021" [Status for progression of B.1.1.7 in Denmark on 17 January 2021]. Statens Serum Institut. Archived from the original on 18 January 2021. Retrieved 19 January 2021.
- "Vietnam reports first case of new coronavirus variant in woman returning from Britain". The Telegraph. 2 January 2021. Archived from the original on 11 January 2022. Retrieved 2 January 2021.
- "New Covid-19 strain found in Luxembourg". luxtimes.lu. Archived from the original on 22 February 2021. Retrieved 4 January 2021.
- "Greece detects four cases of new coronavirus variant". Reuters. 3 January 2021.
- "Four cases of COVID-19 variant confirmed in Jamaica". 3 January 2021. Archived from the original on 3 January 2021. Retrieved 22 January 2021.
- Turner, Katy. "Coronavirus: New, fast-spreading British variant found in Cyprus | Cyprus Mail". Retrieved 4 January 2021.
- "Six cases at NZ border have had new Covid-19 variant, 19 cases in total in the past three days". TVNZ. Retrieved 4 January 2021.
- The Thaiger (3 January 2021). "Britons arriving in Thailand test positive for Covid UK variant". The Thaiger. Retrieved 4 January 2021.
- "Georgia confirms first case of UK-linked coronavirus strain". 4 January 2021.
- "British, South African corornavirus [sic] mutations detected in Austria". Reuters. Berlin. 4 January 2021. Retrieved 6 January 2021.
- "Iran confirms first case of new Covid-19 variant". France 24. Tehran. 5 January 2021. Retrieved 6 January 2021.
- "Oman registers first case of new virus variant in traveller from UK". Reuters. Dubai. 5 January 2021. Retrieved 6 January 2021.
- "Cases of the new UK coronavirus variant have been confirmed in Slovakia". Expats.cz. 5 January 2021.
- "Romania detects first case of British coronavirus variant". Reuters. 8 January 2021.
- "Peru Confirms First Case of COVID-19 Variant Strain". 9 January 2021.
- Mexico detects first case of new coronavirus strain first seen in UK, 11 January 2021 www.business-standard.com, accessed 15 January 2021
- "UK-variant of Covid-19 has reached Malaysia, Dr Noor Hisham confirms". Malay Mail. 11 January 2021.
- "New Covid-19 variant found in Latvia". Baltic News Network. 4 January 2021.
- "Ecuador records first case of new coronavirus variant". 12 January 2021.
- "LIVE UPDATES: COVID-19 pandemic". CNN Philippines. 13 January 2021. Archived from the original on 21 November 2021. Retrieved 15 January 2021.
- "Hungary detects UK variant of coronavirus, surgeon general says". Reuters. 13 January 2021.
- "Gambia records first two cases of British COVID-19 variant". Reuters. 14 January 2021.
- "New COVID-19 strain confirmed to arrive in Dominican Republic from London". 15 January 2021.
- "Argentina detects first case of British virus variant". MedicalXpress. 16 January 2021.
- "UPDATE 1-Czech Republic detects UK coronavirus variant, to maintain lockdown measures". Reuters. 18 January 2021.
- "Kuwait, Morocco report first cases of UK coronavirus variant". The Arab Weekly. 19 January 2021.
- "COVID-19: UK, South Africa variants found in Ghana – Senior Researcher". StarrFM. 19 January 2021. Retrieved 12 February 2021.
- "Kuwait registers first cases of new virus variant". Reuters. 19 January 2021.
- Adebowale, Nike (25 January 2021). "Updated: COVID-19 variant, causing anxiety in UK, found in Nigeria – Official". The Premium Times. Retrieved 29 January 2021.
- "UPDATE 1-Senegal confirms presence of UK variant of coronavirus". Reuters. 28 January 2021.
- "Municipality to test 60,000 residents for British strain of coronavirus". DutchNews. 11 January 2021. Retrieved 12 January 2021.
- Tróndur Olsen (24 January 2021). "UK Covid strain confirmed in the Faroes". Kringvarp Føroya. Retrieved 25 March 2021.
- "North Macedonia reports first case of British coronavirus variant". Reuters. 28 January 2021.
- "Detectaron la variante británica del virus SARS-CoV-2 en Uruguay" (in Spanish). 4 February 2021.
- Telegram.hr. "Potvrđena prva tri slučaja britanskog soja koronavirusa u Hrvatskoj". Telegram.hr (in Croatian). Retrieved 10 February 2021.
- "UK COVID strain found in Sri Lanka - Dr. Chandima Jeewandara". Sri Lanka News - Newsfirst. 12 February 2021. Retrieved 12 February 2021.
- "Coronavirus variant puts N.L. back in lockdown; in-person voting suspended". CBC News. Retrieved 13 February 2021.
- "Menkes Sebut 2 Kasus Mutasi Corona Inggris Masuk dari Saudi". CNN Indonesia (in Indonesian). 2 March 2021. Retrieved 8 March 2021.
- Tunisia records first cases of UK variant – as it happened 2 March 2021 www.theguardian.com, accessed 3 March 2021
- "Covid-19: le variant anglais détecté en Côte d'Ivoire (ministère de la Santé)". Educarriere.ci. 25 March 2021. Retrieved 9 April 2021.
- ^ "Expert reaction new restrictions and the new SARS-CoV-2 variant". Science Media Centre. 21 December 2020. Retrieved 21 December 2020.
Dr Julian Tang, Honorary Associate Professor/Clinical Virologist, University of Leicester, said: "An examination of the global GISAID SARS-COV-2 sequence database shows that this N501Y mutation was actually circulating, sporadically, much earlier in the year outside the UK: in Australia in June–July, USA in July and in Brazil in April, 2020."
- Global Report B.1.351 (Report). cov-lineages.org. 10 March 2021. Retrieved 10 March 2021.
- Tegally H, Wilkinson E, et al. (22 December 2020). "Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa". medRxiv 10.1101/2020.12.21.20248640.
- Global Report P.1 (Report). cov-lineages.org. 11 March 2021. Retrieved 11 March 2021.
- Nuno R. Faria; et al. (12 January 2021). "Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings". Virological.org. Retrieved 11 March 2021.
- "GISAID - hCov19 Variants". www.gisaid.org. Retrieved 13 August 2021.
- "Tracking variants of the Novel Coronavirus in Canada". CTV News. 4 February 2021. Retrieved 13 August 2021.
- "Situation update on coronavirus - Coronavirus variants". Finnish Institute for Health and Welfare. 11 August 2021. Retrieved 11 August 2021.
Alfa-variant: 7,953
- Chand, Meera; Hopkins, Susan; Dabrera, Gavin; Achison, Christina; Barclay, Wendy; Ferguson, Neil; et al. (21 December 2020). Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01 (PDF) (Report). Public Health England. p. 2. Retrieved 22 December 2020.
 This article incorporates text published under the British Open Government Licence v3.0:
This article incorporates text published under the British Open Government Licence v3.0:
- Grabowski, Frederic; Preibisch, Grzegorz; Giziński, Stanisław; Kochańczyk, Marek; Lipniacki, Tomasz (March 2021). "SARS-CoV-2 Variant of Concern 202012/01 Has about Twofold Replicative Advantage and Acquires Concerning Mutations". Viruses. 13 (3): 392. doi:10.3390/v13030392. PMC 8000749. PMID 33804556.
- "Impact of the 69-70del mutation in the spike protein of SARS-CoV-2 on the TaqPath COVID-19 Combo Kit" (PDF). Thermo Fisher Scientific. Retrieved 13 February 2021.
- Dr. Andrea Thorn: The new mutation of SARS-CoV-2 insidecorona.net, accessed 7 February 2021
- "Report 42 - Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data". Imperial College London. Retrieved 26 March 2021.
- "New coronavirus variant: What do we know?". BBC News. 20 December 2020. Retrieved 22 December 2020.
- Ellyatt, Holly (11 February 2021). "UK coronavirus variant on course 'to sweep the world,' leading scientist says". CNBC. Retrieved 26 March 2021.
- "Return to full lockdown might not be enough to control new Covid variant, Sage warns". The Independent. 31 December 2020. Retrieved 31 December 2020.
- "Covid-19: Christmas rules tightened for England, Scotland and Wales". BBC News. 20 December 2020. Retrieved 20 December 2020.
- Fairnie, Robert (19 December 2020). "Travel between Scotland and rest of UK banned over Christmas as border is closed". edinburghlive. Retrieved 20 December 2020.
- ^ Halliday, Josh (21 December 2020). "Calls for national lockdown in England to curb spread of new Covid strain". The Guardian.
- Henley, Jon; Jones, Sam; Giuffrida, Angela; Holmes, Oliver (20 December 2020). "EU to hold crisis talks as countries block travel from UK over new Covid strain". The Guardian.
- Michaels, Daniel (20 December 2020). "Countries Ban Travel From U.K. in Race to Block New Covid-19 Strain". WSJ.
- "Kent lorry queue down to 60 vehicles after border closure". www.bbc.co.uk. 29 December 2020. Retrieved 30 December 2020.
- GRIESHABER, KIRSTEN; HUI, SYLVIA (21 December 2020). "More EU nations ban travel from UK, fearing virus variant". AP NEWS.
- Berger, Miriam (20 December 2020). "Countries across Europe halt flights from Britain over concerns about coronavirus mutation". The Washington Post. ISSN 0190-8286. Archived from the original on 20 December 2020. Retrieved 20 December 2020.
- Quinn, Edna Mohamed(now) Ben; Davies (earlier), Caroline; Davidson, Helen; Wahlquist (earlier), Calla; Walker, Shaun (20 December 2020). "Cases of new strain reported outside of UK – as it happened". The Guardian. ISSN 0261-3077. Retrieved 21 December 2020.
- "Covid-19: UK isolation grows as more countries ban travel". BBC News. 21 December 2020.
- Ogura, Junko. "Japan will ban entry to foreign nationals after Covid-19 variant detected in country". CNN. Retrieved 27 December 2020.
- "California has nation's 2nd confirmed case of virus variant". AP NEWS. 30 December 2020. Retrieved 3 January 2021.
- Mallapaty, Smriti (22 December 2020). "What the data say about border closures and COVID spread". Nature. 589 (7841): 185. doi:10.1038/d41586-020-03605-6. PMID 33361805. S2CID 229692296.
- "COVID-19: 'Delta is making other coronavirus variants extinct' - but what does it mean for the UK?". Sky News. Retrieved 17 August 2022.
- Rachael Rettner (23 March 2022). "Coronavirus variants: Facts about omicron, delta and other COVID-19 mutants". livescience.com. Retrieved 17 August 2022.
- "UK reports new variant, termed VUI 202012/01". GISAID. Archived from the original on 20 December 2020. Retrieved 20 December 2020.
- Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom (PDF) (Report). European Centre for Disease Prevention and Control (ECDC). 20 December 2020.
External links
- Corum, Jonathan; Zimmer, Carl (18 January 2021). "Inside the B.1.1.7 Coronavirus Variant". The New York Times.
- Public Health England: Variants: UK distribution – summary of 4 nations distribution
- PANGO lineages: New Variant Report - Report on global distribution of lineage B.1.1.7
- GISAID sequencing data base - Tracking of COVID-19 variants
| Pandemics, epidemics and notable disease outbreaks | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Local |
| ||||||||||||||||||||
| Global |
| ||||||||||||||||||||
| Taxonomy of the Coronaviridae | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Higher taxonomy: Riboviria > Orthornavirae > Pisuviricota > Pisoniviricetes > Nidovirales > Cornidovirineae > Coronaviridae | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: ICTV –– | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| COVID-19 pandemic | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 every two weeks. Another group came to similar conclusions, generating a replicative advantage, independent of "protective measures", of 2.24 per generation of 6.73 days, out-competing the ancestral variant by
every two weeks. Another group came to similar conclusions, generating a replicative advantage, independent of "protective measures", of 2.24 per generation of 6.73 days, out-competing the ancestral variant by  every two weeks. Similarly, in Ireland, the variant—as indicated by the missing
every two weeks. Similarly, in Ireland, the variant—as indicated by the missing  , when compared to the average growth for all other variants by the end of this two week period. The variant became the dominant variant in London, East of England and the South East from low levels in one to two months. A surge of SARS-CoV-2 infections around the start of the new year is seen as being the result of the elevated transmissibility of the variant, while the other variants were in decline.
, when compared to the average growth for all other variants by the end of this two week period. The variant became the dominant variant in London, East of England and the South East from low levels in one to two months. A surge of SARS-CoV-2 infections around the start of the new year is seen as being the result of the elevated transmissibility of the variant, while the other variants were in decline.