| Betalipothrixvirus | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Adnaviria |
| Kingdom: | Zilligvirae |
| Phylum: | Taleaviricota |
| Class: | Tokiviricetes |
| Order: | Ligamenvirales |
| Family: | Lipothrixviridae |
| Genus: | Betalipothrixvirus |
| Species | |
| |
Betalipothrixvirus is a genus of viruses in the family Lipothrixviridae. Archaea serve as natural hosts. The genus contains six species.
All species within the genus have linear double-stranded DNA genomes and some infect hyperthermophilic crenarchaea of the orders Sulfolobales.
Etymology: the root of the word (Beta)Lipothrix: from Greek, "fat hair", referring to the virion shape(SIB).
Taxonomy
The following six species are assigned to the genus:
- Betalipothrixvirus acidiani
- Betalipothrixvirus hveragerdiense
- Betalipothrixvirus pezzuloense
- Betalipothrixvirus pozzuoliense
- Betalipothrixvirus puteoliense
- Betalipothrixvirus uzonense
They were previously known as (in order):
- Acidianus filamentous virus 3 - AFV3
- Sulfolobus islandicus filamentous virus - SIFV
- Acidianus filamentous virus 7 - AFV7
- Acidianus filamentous virus 6 - AFV6
- Acidianus filamentous virus 8 - AFV8
- Acidianus filamentous virus 9 - AFV9
Structure
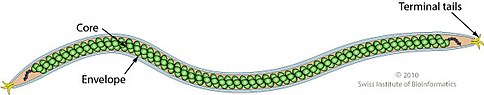
Viruses in Betalipothrixvirus are enveloped, with rod-shaped geometries. The diameter is around 24 nm, with a length of 2000 nm. Genomes are linear, around 40.5kb in length. The genome has 67 open reading frames.
It is important to note that there are similarities between the structure of the species within the genus. For example, the core of the SIFV species is formed by “a zipper-like array of DNA-associated protein subunits and is covered by a lipid envelope containing host lipids.” Similarly, in images of the structure of AFV3, zipper or zigzag patterned high-density “ovoid masses” have been found inside the lumen of the virus. This is likely the projection of a double row of nucleosome-like particles.
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Betalipothrixvirus | Rod-shaped | Enveloped | Linear | Monopartite |

Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by adsorption into the host cell. DNA templated transcription is the method of transcription. Archaea serve as the natural host. Transmission routes are passive diffusion.
It is important to note that all six members of the genus Betalipothrixvirus have demonstrated a high level of conservation in both gene content and gene order over large regions. These large regions have been predicted to produce a putative helicase, a nuclease, a protein phosphatase, transcription regulators and glycosyltransferases. Furthermore, studies have shown that some of the shared features of the genomes of these six species include (1) large inverted terminal repeats exhibiting conserved, regularly spaced direct repeats; (2) a highly conserved operon encoding the two major structural proteins; (3) multiple overlapping open reading frames, which may be indicative of gene recoding; (4) putative 12-bp genetic elements; and (5) partial gene sequences corresponding closely to spacer sequences of chromosomal repeat clusters. Additionally, the existing wikipedia page states that “The genome has 67 open reading frames''. I believe it is important to add and clarify that there have been 58 to 67 open reading frames (ORFs) (>48 amino acids) identified for each virus. Finally, the table included on the wikipedia page does not indicate the type of symmetry of the virus, and is left blank. Since it is known that the shape of the virus is rod-like, the symmetry is helical, and therefore the blank in the table can be filled.
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Betalipothrixvirus | Archea | None | Injection | Budding | Cytoplasm | Cytoplasm | Passive diffusion |
References
- ^ "Viral Zone". ExPASy. Retrieved 15 June 2015.
- ^ "Virus Taxonomy: 2023 Release". International Committee on Taxonomy of Viruses (ICTV). April 2024. Retrieved 21 May 2024.
- Prangishvili, D.; Garrett, R.A. (1 April 2004). "Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses". Biochemical Society Transactions. 32 (2): 204–208. doi:10.1042/bst0320204. ISSN 0300-5127.
- Arnold, Hans Peter; Zillig, Wolfram; Ziese, Ulrike; Holz, Ingelore; Crosby, Marie; Utterback, Terry; Weidmann, Jan F; Kristjanson, Jakob K; Klenk, Hans Peter; Nelson, Karen E; Fraser, Claire M (February 2000). "A Novel Lipothrixvirus, SIFV, of the Extremely Thermophilic Crenarchaeon Sulfolobus". Virology. 267 (2): 252–266. doi:10.1006/viro.1999.0105.
- Vestergaard, Gisle; Aramayo, Ricardo; Basta, Tamara; Häring, Monika; Peng, Xu; Brügger, Kim; Chen, Lanming; Rachel, Reinhard; Boisset, Nicolas; Garrett, Roger A.; Prangishvili, David (January 2008). "Structure of the Acidianus Filamentous Virus 3 and Comparative Genomics of Related Archaeal Lipothrixviruses". Journal of Virology. 82 (1): 371–381. doi:10.1128/JVI.01410-07. ISSN 0022-538X. PMC 2224351. PMID 17942536.
- Vestergaard, Gisle; Aramayo, Ricardo; Basta, Tamara; Häring, Monika; Peng, Xu; Brügger, Kim; Chen, Lanming; Rachel, Reinhard; Boisset, Nicolas; Garrett, Roger A.; Prangishvili, David (January 2008). "Structure of the Acidianus Filamentous Virus 3 and Comparative Genomics of Related Archaeal Lipothrixviruses". Journal of Virology. 82 (1): 371–381. doi:10.1128/JVI.01410-07. ISSN 0022-538X. PMC 2224351. PMID 17942536.
- Vestergaard, Gisle; Aramayo, Ricardo; Basta, Tamara; Häring, Monika; Peng, Xu; Brügger, Kim; Chen, Lanming; Rachel, Reinhard; Boisset, Nicolas; Garrett, Roger A.; Prangishvili, David (January 2008). "Structure of the Acidianus Filamentous Virus 3 and Comparative Genomics of Related Archaeal Lipothrixviruses". Journal of Virology. 82 (1): 371–381. doi:10.1128/JVI.01410-07. ISSN 0022-538X. PMC 2224351. PMID 17942536.
- Vestergaard, Gisle; Aramayo, Ricardo; Basta, Tamara; Häring, Monika; Peng, Xu; Brügger, Kim; Chen, Lanming; Rachel, Reinhard; Boisset, Nicolas; Garrett, Roger A.; Prangishvili, David (January 2008). "Structure of the Acidianus Filamentous Virus 3 and Comparative Genomics of Related Archaeal Lipothrixviruses". Journal of Virology. 82 (1): 371–381. doi:10.1128/JVI.01410-07. ISSN 0022-538X. PMC 2224351. PMID 17942536.
- Johnson, J.E.; Speir, J.A. (2008), "Virus Particle Structure: Principles", Encyclopedia of Virology, Elsevier, pp. 393–401, doi:10.1016/b978-012374410-4.00598-7, ISBN 978-0-12-374410-4, retrieved 6 May 2023
Notes
- On the left is their previous species name and their virus name, on the right is their virus name abbreviation
External links
| Taxon identifiers | |
|---|---|
| Betalipothrixvirus | |