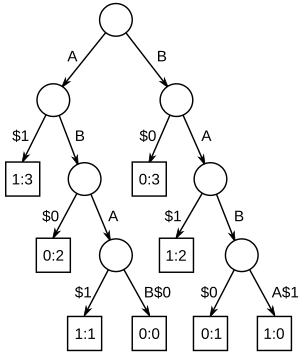
ABAB and BABA. Suffix links not shown.In computer science, a generalized suffix tree is a suffix tree for a set of strings. Given the set of strings of total length , it is a Patricia tree containing all suffixes of the strings. It is mostly used in bioinformatics.
Functionality
It can be built in time and space, and can be used to find all occurrences of a string of length in time, which is asymptotically optimal (assuming the size of the alphabet is constant).
When constructing such a tree, each string should be padded with a unique out-of-alphabet marker symbol (or string) to ensure no suffix is a substring of another, guaranteeing each suffix is represented by a unique leaf node.
Algorithms for constructing a GST include Ukkonen's algorithm (1995) and McCreight's algorithm (1976).
Example
A suffix tree for the strings ABAB and BABA is shown in a figure above. They are padded with the unique terminator strings $0 and $1. The numbers in the leaf nodes are string number and starting position. Notice how a left to right traversal of the leaf nodes corresponds to the sorted order of the suffixes. The terminators might be strings or unique single symbols. Edges on $ from the root are left out in this example.
Alternatives
An alternative to building a generalized suffix tree is to concatenate the strings, and build a regular suffix tree or suffix array for the resulting string. When hits are evaluated after a search, global positions are mapped into documents and local positions with some algorithm and/or data structure, such as a binary search in the starting/ending positions of the documents.
References
- Paul Bieganski; John Riedl; John Carlis; Ernest F. Retzel (1994). "Generalized Suffix Trees for Biological Sequence Data". Biotechnology Computing, Proceedings of the Twenty-Seventh Hawaii International Conference on. pp. 35–44. doi:10.1109/HICSS.1994.323593.
- Gusfield, Dan (1999) . Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. USA: Cambridge University Press. ISBN 978-0-521-58519-4.
External links
 Media related to Generalized suffix tree at Wikimedia Commons
Media related to Generalized suffix tree at Wikimedia Commons- A C implementation of Generalized Suffix Tree for two strings
| Strings | |
|---|---|
| String metric | |
| String-searching algorithm | |
| Multiple string searching | |
| Regular expression | |
| Sequence alignment | |
| Data structure | |
| Other | |
 of total length
of total length  , it is a
, it is a  time and space, and can be used to find all
time and space, and can be used to find all  occurrences of a string
occurrences of a string  of length
of length  in
in  time, which is
time, which is