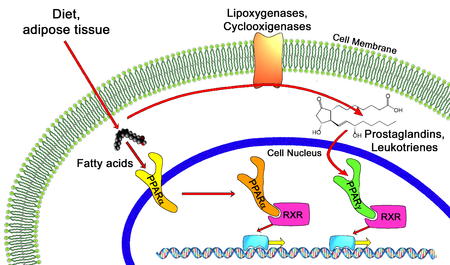
In the field of molecular biology, the peroxisome proliferator–activated receptors (PPARs) are a group of nuclear receptor proteins that function as transcription factors regulating the expression of genes. PPARs play essential roles in the regulation of cellular differentiation, development, and metabolism (carbohydrate, lipid, protein), and tumorigenesis of higher organisms.
Nomenclature and tissue distribution
| Peroxisome proliferator-activated receptor alpha | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | PPARA | ||||||
| Alt. symbols | PPAR | ||||||
| NCBI gene | 5465 | ||||||
| HGNC | 9232 | ||||||
| OMIM | 170998 | ||||||
| RefSeq | NM_001001928 | ||||||
| UniProt | Q07869 | ||||||
| Other data | |||||||
| Locus | Chr. 22 q12-q13.1 | ||||||
| |||||||
| Peroxisome proliferator-activated receptor gamma | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Symbol | PPARG | ||||||
| NCBI gene | 5468 | ||||||
| HGNC | 9236 | ||||||
| OMIM | 601487 | ||||||
| RefSeq | NM_005037 | ||||||
| UniProt | P37231 | ||||||
| Other data | |||||||
| Locus | Chr. 3 p25 | ||||||
| |||||||
| Peroxisome proliferator-activated receptor delta | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | PPARD | ||||||
| NCBI gene | 5467 | ||||||
| HGNC | 9235 | ||||||
| OMIM | 600409 | ||||||
| RefSeq | NM_006238 | ||||||
| UniProt | Q03181 | ||||||
| Other data | |||||||
| Locus | Chr. 6 p21.2 | ||||||
| |||||||
Three types of PPARs have been identified: alpha, gamma, and delta (beta):
- α (alpha) - expressed in liver, kidney, heart, muscle, adipose tissue, and others
- β/δ (beta/delta) - expressed in many tissues, especially in brain, adipose tissue, and skin
- γ (gamma) - although transcribed by the same gene, this PPAR, by way of alternative splicing, is expressed in three forms:
- γ1 - expressed in virtually all tissues, including heart, muscle, colon, kidney, pancreas, and spleen
- γ2 - expressed mainly in adipose tissue; it is 30 amino acids longer than γ1
- γ3 - expressed in macrophages, large intestine, white adipose tissue
History
These agents, pharmacologically related to the fibrates were discovered in the early 1980s.
PPARs were originally identified in Xenopus frogs as receptors that induce the proliferation of peroxisomes in cells in 1992. The first PPAR (PPARα) was discovered in 1990 during the search for a molecular target of a group of agents then referred to as peroxisome proliferators, as they increased peroxisomal numbers in rodent liver tissue, apart from improving insulin sensitivity.
When it turned out that PPARs played a much more versatile role in biology, the agents were in turn termed PPAR ligands. The best-known PPAR ligands are the thiazolidinediones.
After PPARδ (delta) was identified in humans in 1992, it turned out to be closely related to PPARβ (beta), previously described during the same year in an amphibian, Xenopus. The term "PPARδ" is generally used in the US, whereas the use of "PPARβ" has remained in Europe, where this receptor was initially discovered in Xenopus.
PPARs were so-named because they were discovered to induce peroxisome proliferation in rodents, but this induction of peroxisome proliferation is not believed to occur in humans.
Physiological function
All PPARs heterodimerize with the retinoid X receptor (RXR) and bind to specific regions on the DNA of target genes. These DNA sequences are termed PPREs (peroxisome proliferator hormone response elements). The DNA consensus sequence is AGGTCANAGGTCA, with N being any nucleotide. In general, this sequence occurs in the promoter region of a gene, and, when the PPAR binds its ligand, transcription of target genes is increased or decreased, depending on the gene. The RXR also forms a heterodimer with a number of other receptors (e.g., vitamin D and thyroid hormone).
The function of PPARs is modified by the precise shape of their ligand-binding domain (see below) induced by ligand binding and by a number of coactivator and corepressor proteins, the presence of which can stimulate or inhibit receptor function, respectively.
Endogenous ligands for the PPARs include free fatty acids, eicosanoids and Vitamin B3. PPARγ is activated by PGJ2 (a prostaglandin) and certain members of the 5-HETE family of arachidonic acid metabolites including 5-oxo-15(S)-HETE and 5-oxo-ETE. In contrast, PPARα is activated by leukotriene B4. Certain members of the 15-hydroxyeicosatetraenoic acid family of arachidonic acid metabolites, including 15(S)-HETE, 15(R)-HETE, and 15-HpETE activate to varying degrees PPAR alpha, beta/delta, and gamma. In addition, PPARγ was reported to be involved in cancer pathogenesis and growth. PPARγ activation by agonist RS5444 may inhibit anaplastic thyroid cancer growth. See for a review and critique of the roles of PPAR gamma in cancer.
Genetics
The three main forms of PPAR are transcribed from different genes:
- PPARα - chromosome 22q12-13.1 (OMIM 170998)
- PPARβ/δ - chromosome 6p21.2-21.1 (OMIM 600409)
- PPARγ - chromosome 3p25 (OMIM 601487).
Hereditary disorders of all 3 of these PPARs have been described, generally leading to a loss in function and concomitant lipodystrophy, insulin resistance, and/or acanthosis nigricans. Of PPARγ, a gain-of-function mutation has been described and studied: Pro12Ala, which decreases the risk of insulin resistance. It is quite prevalent, with an allele frequency of 0.03 - 0.12 in some populations. In contrast, pro115gln is associated with obesity. Certain other polymorphisms in PPAR show a high incidence in populations with elevated body mass indexes.
Structure
Like other nuclear receptors, PPARs are modular in structure and contain the following functional domains:
- (A/B) - N-terminal region
- (C) DBD - DNA-binding domain
- (D) - flexible hinge region
- (E) LBD - ligand binding domain
- (F) C-terminal region
The DBD contains two zinc finger motifs, which bind to specific sequences of DNA known as hormone response elements when the receptor is activated.
The LBD has an extensive secondary structure consisting of 13 alpha helices and a beta sheet. Both natural and synthetic ligands can bind to the LBD, either activating or repressing the receptor's activity.
Pharmacology and PPAR modulators
Main article: PPAR modulatorPPARα and PPARγ are the molecular targets of a number of marketed drugs.
For instance the hypolipidemic fibrates activate PPARα.
The anti diabetic thiazolidinediones activate PPARγ.
The synthetic chemical perfluorooctanoic acid activates PPARα while perfluorononanoic acid activates both PPARα and PPARγ.
Berberine inactivates PPARγ. Huang C, Zhang Y, Gong Z, Sheng X, Li Z, Zhang W, Qin Y (2006). "Berberine inhibits 3T3-L1 adipocyte differentiation through the PPARgamma pathway". Biochemical and Biophysical Research Communications. 348 (2): 571–578. doi:10.1016/j.bbrc.2006.07.095. PMID 16890192.</ref>
Other natural compounds from different chemical classes activate or inactivate PPARγ.
See also
References
- Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O'Rahilly S, Palmer CN, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W (2006). "International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors". Pharmacol. Rev. 58 (4): 726–41. doi:10.1124/pr.58.4.5. PMID 17132851. S2CID 2240461.
- Dunning, Kylie R.; Anastasi, Marie R.; Zhang, Voueleng J.; Russell, Darryl L.; Robker, Rebecca L. (2014-02-05). "Regulation of Fatty Acid Oxidation in Mouse Cumulus-Oocyte Complexes during Maturation and Modulation by PPAR Agonists". PLOS ONE. 9 (2): e87327. Bibcode:2014PLoSO...987327D. doi:10.1371/journal.pone.0087327. ISSN 1932-6203. PMC 3914821. PMID 24505284.
- Belfiore A, Genua M, Malaguarnera R (2009). "PPAR-gamma Agonists and Their Effects on IGF-I Receptor Signaling: Implications for Cancer". PPAR Res. 2009: 830501. doi:10.1155/2009/830501. PMC 2709717. PMID 19609453.
- ^ Berger J, Moller DE (2002). "The mechanisms of action of PPARs". Annu. Rev. Med. 53: 409–35. doi:10.1146/annurev.med.53.082901.104018. PMID 11818483.
- Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W (2006). "From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions". Prog. Lipid Res. 45 (2): 120–59. doi:10.1016/j.plipres.2005.12.002. PMID 16476485.
- Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S (October 2011). "The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases". J Adv Pharm Technol Res. 2 (4): 236–40. doi:10.4103/2231-4040.90879. PMC 3255347. PMID 22247890.
- Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W (1992). "Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors". Cell. 68 (5): 879–87. doi:10.1016/0092-8674(92)90031-7. PMID 1312391. S2CID 3148132.
- Issemann I, Green S (1990). "Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators". Nature. 347 (6294): 645–50. Bibcode:1990Natur.347..645I. doi:10.1038/347645a0. PMID 2129546. S2CID 4306126.
- Schmidt A, Endo N, Rutledge SJ, Vogel R, Shinar D, Rodan GA (1992). "Identification of a new member of the steroid hormone receptor superfamily that is activated by a peroxisome proliferator and fatty acids". Mol. Endocrinol. 6 (10): 1634–41. doi:10.1210/mend.6.10.1333051. PMID 1333051. S2CID 23506853.
- Corton JC, Peters JM, Klaunig JE (2018). "The PPARα-dependent rodent liver tumor response is not relevant to humans: addressing misconceptions". Journal of Molecular Endocrinology. 92 (1): 83–119. Bibcode:2018ArTox..92...83C. doi:10.1007/s00204-017-2094-7. PMC 6092738. PMID 29197930.
- Sugden MC, Caton PW, Holness MJ, Miller JJ (2021). "Peroxisome Proliferator-Activated Receptors". Reference Module in Life Sciences. Vol. 17. Elsevier. pp. 574–583. doi:10.1016/B978-0-12-819460-7.00200-0. ISBN 9780128096338. S2CID 241510571.
- Yu S, Reddy JK (2007). "Transcription coactivators for peroxisome proliferator-activated receptors". Biochim. Biophys. Acta. 1771 (8): 936–51. doi:10.1016/j.bbalip.2007.01.008. PMID 17306620.
- Biochim. Biophys. Acta 1736:228–236, 2005
- Ezzeddini R, Taghikhani M, Salek Farrokhi A, Somi MH, Samadi N, Esfahani A, Rasaee, MJ (May 2021). "Downregulation of fatty acid oxidation by involvement of HIF-1α and PPARγ in human gastric adenocarcinoma and its related clinical significance". Journal of Physiology and Biochemistry. 77 (2): 249–260. doi:10.1007/s13105-021-00791-3. PMID 33730333. S2CID 232300877.
- Mol. Pharmacol. 77-171-184, 2010
- Marlow LA, Reynolds LA, Cleland AS, Cooper SJ, Gumz ML, Kurakata S, Fujiwara K, Zhang Y, Sebo T, Grant C, McIver B, Wadsworth JT, Radisky DC, Smallridge RC, Copland JA (February 2009). "Reactivation of suppressed RhoB is a critical step for the inhibition of anaplastic thyroid cancer growth". Cancer Res. 69 (4): 1536–44. doi:10.1158/0008-5472.CAN-08-3718. PMC 2644344. PMID 19208833.
- Curr. Mol. Med. 7:532–540, 2007
- Meirhaeghe A, Amouyel P (2004). "Impact of genetic variation of PPARgamma in humans". Mol. Genet. Metab. 83 (1–2): 93–102. doi:10.1016/j.ymgme.2004.08.014. PMID 15464424.
- Buzzetti R, Petrone A, Ribaudo MC, Alemanno I, Zavarella S, Mein CA, Maiani F, Tiberti C, Baroni MG, Vecci E, Arca M, Leonetti F, Di Mario U (2004). "The common PPAR-gamma2 Pro12Ala variant is associated with greater insulin sensitivity". European Journal of Human Genetics. 12 (12): 1050–4. doi:10.1038/sj.ejhg.5201283. PMID 15367918.
- Zoete V, Grosdidier A, Michielin O (2007). "Peroxisome proliferator-activated receptor structures: ligand specificity, molecular switch and interactions with regulators". Biochim. Biophys. Acta. 1771 (8): 915–25. doi:10.1016/j.bbalip.2007.01.007. PMID 17317294.
- Atanasov AG, Wang JN, Gu SP, Bu J, Kramer MP, Baumgartner L, Fakhrudin N, Ladurner A, Malainer C, Vuorinen A, Noha SM, Schwaiger S, Rollinger JM, Schuster D, Stuppner H, Dirsch VM, Heiss EH (2013). "Honokiol: a non-adipogenic PPARγ agonist from nature". Biochim. Biophys. Acta. 1830 (10): 4813–9. doi:10.1016/j.bbagen.2013.06.021. PMC 3790966. PMID 23811337.
- Atanasov AG, Blunder M, Fakhrudin N, Liu X, Noha SM, Malainer C, Kramer MP, Cocic A, Kunert O, Schinkovitz A, Heiss EH, Schuster D, Dirsch VM, Bauer R (2013). "Polyacetylenes from Notopterygium incisum--new selective partial agonists of peroxisome proliferator-activated receptor-gamma". PLOS ONE. 8 (4): e61755. Bibcode:2013PLoSO...861755A. doi:10.1371/journal.pone.0061755. PMC 3632601. PMID 23630612.
- Ammazzalorso, Alessandra; Amoroso, Rosa (2019-02-28). "Inhibition of PPARγ by Natural Compounds as a Promising Strategy in Obesity and Diabetes". The Open Medicinal Chemistry Journal. 13 (1): 7–15. doi:10.2174/1874104501913010007.
External links
- (PPAR Resource Page, Penn State University).
- (Nuclear Receptor Resource).
- PPAR reference outline Archived 2020-02-14 at the Wayback Machine (Rutgers University).
- Peroxisome+Proliferator-Activated+Receptors at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Proteopedia Peroxisome_Proliferator-Activated_Receptors - the Peroxisome Proliferator-Activated Receptor Structure in Interactive 3D
| Transcription factors and intracellular receptors | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| see also transcription factor/coregulator deficiencies | |||||||||||||||||||||||||||||||