| |
| |

| |
| Names | |
|---|---|
| IUPAC name 9H-purine | |
| Identifiers | |
| CAS Number | |
| 3D model (JSmol) | |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.004.020 |
| KEGG | |
| MeSH | Purine |
| PubChem CID | |
| UNII | |
| CompTox Dashboard (EPA) | |
InChI
| |
SMILES
| |
| Properties | |
| Chemical formula | C5H4N4 |
| Molar mass | 120.115 g·mol |
| Melting point | 214 °C (417 °F; 487 K) |
| Solubility in water | 500 g/L (RT) |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C , 100 kPa).
| |
Purine is a heterocyclic aromatic organic compound that consists of two rings (pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature.
Dietary sources
Purines are found in high concentration in meat and meat products, especially internal organs such as liver and kidney. In general, plant-based diets are low in purines. High-purine plants and algae include some legumes (lentils, soybeans, and black-eyed peas) and spirulina. Examples of high-purine sources include: sweetbreads, anchovies, sardines, liver, beef kidneys, brains, meat extracts (e.g., Oxo, Bovril), herring, mackerel, scallops, game meats, yeast (beer, yeast extract, nutritional yeast) and gravy.
A moderate amount of purine is also contained in red meat, beef, pork, poultry, fish and seafood, asparagus, cauliflower, spinach, mushrooms, green peas, lentils, dried peas, beans, oatmeal, wheat bran, wheat germ, and haws.
Biochemistry
Purines and pyrimidines make up the two groups of nitrogenous bases, including the two groups of nucleotide bases. The purine bases are guanine (G) and adenine (A) which form corresponding nucleosides-deoxyribonucleosides (deoxyguanosine and deoxyadenosine) with deoxyribose moiety and ribonucleosides (guanosine, adenosine) with ribose moiety. These nucleosides with phosphoric acid form corresponding nucleotides (deoxyguanylate, deoxyadenylate and guanylate, adenylate) which are the building blocks of DNA and RNA, respectively. Purine bases also play an essential role in many metabolic and signalling processes within the compounds guanosine monophosphate (GMP) and adenosine monophosphate (AMP).
In order to perform these essential cellular processes, both purines and pyrimidines are needed by the cell, and in similar quantities. Both purine and pyrimidine are self-inhibiting and activating. When purines are formed, they inhibit the enzymes required for more purine formation. This self-inhibition occurs as they also activate the enzymes needed for pyrimidine formation. Pyrimidine simultaneously self-inhibits and activates purine in a similar manner. Because of this, there is nearly an equal amount of both substances in the cell at all times.
Properties
Purine is both a very weak acid (pKa 8.93) and an even weaker base (pKa 2.39). If dissolved in pure water, the pH is halfway between these two pKa values.
Purine is aromatic, having four tautomers each with a hydrogen bonded to a different one of the four nitrogen atoms. These are identified as 1-H, 3-H, 7-H, and 9-H (see image of numbered ring). The common crystalline form favours the 7-H tautomer, while in polar solvents both the 9-H and 7-H tautomers predominate. Substituents to the rings and interactions with other molecules can shift the equilibrium of these tautomers.
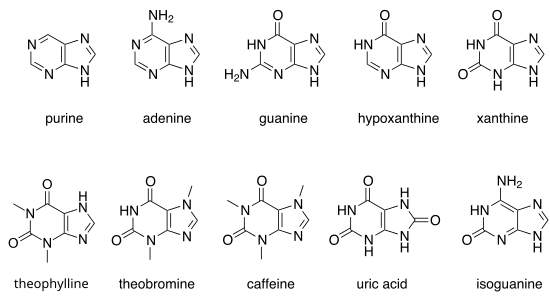
Notable purines
There are many naturally occurring purines. They include the nucleotide bases adenine and guanine. In DNA, these bases form hydrogen bonds with their complementary pyrimidines, thymine and cytosine, respectively. This is called complementary base pairing. In RNA, the complement of adenine is uracil instead of thymine.
Other notable purines are hypoxanthine, xanthine, theophylline, theobromine, caffeine, uric acid and isoguanine.
Functions
Aside from the crucial roles of purines (adenine and guanine) in DNA and RNA, purines are also significant components in a number of other important biomolecules, such as ATP, GTP, cyclic AMP, NADH, and coenzyme A. Purine (1) itself, has not been found in nature, but it can be produced by organic synthesis.
They may also function directly as neurotransmitters, acting upon purinergic receptors. Adenosine activates adenosine receptors.
History
The word purine (pure urine) was coined by the German chemist Emil Fischer in 1884. He synthesized it for the first time in 1898. The starting material for the reaction sequence was uric acid (8), which had been isolated from kidney stones by Carl Wilhelm Scheele in 1776. Uric acid was reacted with PCl5 to give 2,6,8-trichloropurine, which was converted with HI and PH4I to give 2,6-diiodopurine. The product was reduced to purine using zinc dust.

Conversion of uric acid (left) to purine (right) via 2,6,8-trichloropurine and 2,6-diiodopurine intermediates
Metabolism
Main article: Purine metabolismMany organisms have metabolic pathways to synthesize and break down purines.
Purines are biologically synthesized as nucleosides (bases attached to ribose).
Accumulation of modified purine nucleotides is defective to various cellular processes, especially those involving DNA and RNA. To be viable, organisms possess a number of deoxypurine phosphohydrolases, which hydrolyze these purine derivatives removing them from the active NTP and dNTP pools. Deamination of purine bases can result in accumulation of such nucleotides as ITP, dITP, XTP and dXTP.
Defects in enzymes that control purine production and breakdown can severely alter a cell's DNA sequences, which may explain why people who carry certain genetic variants of purine metabolic enzymes have a higher risk for some types of cancer.
Purine biosynthesis in the three domains of life
Organisms in all three domains of life, eukaryotes, bacteria and archaea, are able to carry out de novo biosynthesis of purines. This ability reflects the essentiality of purines for life. The biochemical pathway of synthesis is very similar in eukaryotes and bacterial species, but is more variable among archaeal species. A nearly complete, or complete, set of genes required for purine biosynthesis was determined to be present in 58 of the 65 archaeal species studied. However, also identified were seven archaeal species with entirely, or nearly entirely, absent purine encoding genes. Apparently the archaeal species unable to synthesize purines are able to acquire exogenous purines for growth., and are thus analogous to purine mutants of eukaryotes, e.g. purine mutants of the Ascomycete fungus Neurospora crassa, that also require exogenous purines for growth.
Relationship with gout
See also: Gout § CauseHigher levels of meat and seafood consumption are associated with an increased risk of gout, whereas a higher level of consumption of dairy products is associated with a decreased risk. Moderate intake of purine-rich vegetables or protein is not associated with an increased risk of gout. Similar results have been found with the risk of hyperuricemia.
Laboratory synthesis
In addition to in vivo synthesis of purines in purine metabolism, purine can also be synthesized artificially.
Purine is obtained in good yield when formamide is heated in an open vessel at 170 °C for 28 hours.
This remarkable reaction and others like it have been discussed in the context of the origin of life.
Patented August 20, 1968, the current recognized method of industrial-scale production of adenine is a modified form of the formamide method. This method heats up formamide under 120 °C conditions within a sealed flask for 5 hours to form adenine. The reaction is heavily increased in quantity by using a phosphorus oxychloride (phosphoryl chloride) or phosphorus pentachloride as an acid catalyst and sunlight or ultraviolet conditions. After the 5 hours have passed and the formamide-phosphorus oxychloride-adenine solution cools down, water is put into the flask containing the formamide and now-formed adenine. The water-formamide-adenine solution is then poured through a filtering column of activated charcoal. The water and formamide molecules, being small molecules, will pass through the charcoal and into the waste flask; the large adenine molecules, however, will attach or “adsorb” to the charcoal due to the van der waals forces that interact between the adenine and the carbon in the charcoal. Because charcoal has a large surface area, it's able to capture the majority of molecules that pass a certain size (greater than water and formamide) through it. To extract the adenine from the charcoal-adsorbed adenine, ammonia gas dissolved in water (aqua ammonia) is poured onto the activated charcoal-adenine structure to liberate the adenine into the ammonia-water solution. The solution containing water, ammonia, and adenine is then left to air dry, with the adenine losing solubility due to the loss of ammonia gas that previously made the solution basic and capable of dissolving adenine, thus causing it to crystallize into a pure white powder that can be stored.
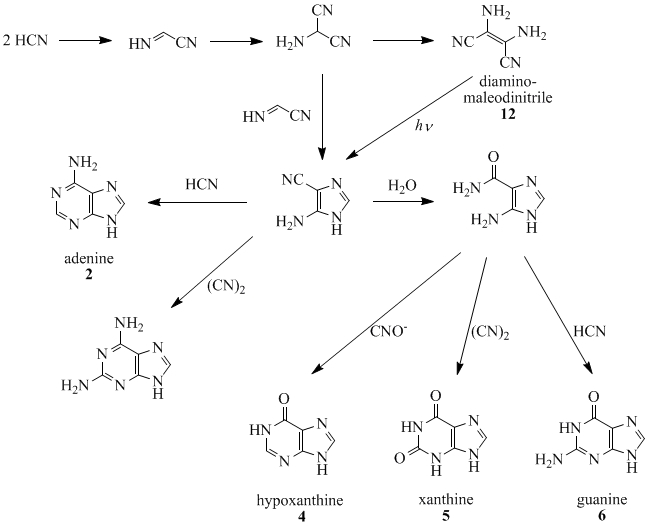
Oro and Kamat (1961) and Orgel co-workers (1966, 1967) have shown that four molecules of HCN tetramerize to form diaminomaleodinitrile (12), which can be converted into almost all naturally occurring purines. For example, five molecules of HCN condense in an exothermic reaction to make adenine, especially in the presence of ammonia.
The Traube purine synthesis (1900) is a classic reaction (named after Wilhelm Traube) between an amine-substituted pyrimidine and formic acid.
Prebiotic synthesis of purine ribonucleosides
In order to understand how life arose, knowledge is required of the chemical pathways that permit formation of the key building blocks of life under plausible prebiotic conditions. Nam et al. (2018) demonstrated the direct condensation of purine and pyrimidine nucleobases with ribose to give ribonucleosides in aqueous microdroplets, a key step leading to RNA formation. Also, a plausible prebiotic process for synthesizing purine ribonucleosides was presented by Becker et al. in 2016.
See also
- Purinones
- Pyrimidine
- Simple aromatic rings
- Transition
- Transversion
- Gout, a disorder of purine metabolism
- Adenine
- Guanine
References
- Rosemeyer H (March 2004). "The chemodiversity of purine as a constituent of natural products". Chemistry & Biodiversity. 1 (3): 361–401. doi:10.1002/cbdv.200490033. PMID 17191854. S2CID 12416667.
- "Gout: List of Foods High and Low in Purine Content". Dietaryfiberfood.com. 2016-04-08. Archived from the original on 2011-11-12. Retrieved 2016-07-16.
- Kaneko K, Aoyagi Y, Fukuuchi T, Inazawa K, Yamaoka N (2014). "Total purine and purine base content of common foodstuffs for facilitating nutritional therapy for gout and hyperuricemia". Biological & Pharmaceutical Bulletin. 37 (5): 709–721. doi:10.1248/bpb.b13-00967. PMID 24553148.
- "Gout Diet: What Foods To Avoid". Healthcastle.com. Archived from the original on 2017-08-14. Retrieved 2016-07-16.
- Guyton AC (2006). Textbook of Medical Physiology. Elsevier. p. 37. ISBN 978-0-7216-0240-0.
- Seela F, et al. (2014). "Hetarenes III (Six-Membered Rings and Larger Hetero-Rings with Maximum Unsaturation) — Part 2b". In Schaumann E (ed.). Houben-Weyl Methods of Organic Chemistry. Vol. E 9b/2 (4th Supplement ed.). Thieme. p. 310. ISBN 978-3-13-181504-0. Archived from the original on 2022-02-17. Retrieved 2020-05-15.
- Raczyńska ED, Gal JF, Maria PC, Kamińska B, Igielska M, Kurpiewski J, Juras W (April 2020). "Purine tautomeric preferences and bond-length alternation in relation with protonation-deprotonation and alkali metal cationization". Journal of Molecular Modeling. 26 (5): 93. doi:10.1007/s00894-020-4343-6. PMC 7256107. PMID 32248379.
- Stasyuk OA, Szatyłowicz H, Krygowski TM (April 2012). "Effect of the H-bonding on aromaticity of purine tautomers". The Journal of Organic Chemistry. 77 (8): 4035–45. doi:10.1021/jo300406r. PMID 22448684.
- McGuigan H (1921). An Introduction To Chemical Pharmacology. P. Blakiston's Sons & Co. p. 283. Archived from the original on April 16, 2020. Retrieved July 18, 2012.
- Fischer E (1884). "Ueber die Harnsäure. I." [On uric acid. I.] (PDF). Berichte der Deutschen Chemischen Gesellschaft. 17: 328–338. doi:10.1002/cber.18840170196. Retrieved 2016-04-20.

From p. 329 Archived 2022-02-17 at the Wayback Machine: "Um eine rationelle Nomenklatur der so entstehenden zahlreichen Substanzen zu ermöglichen, betrachte ich dieselben als Abkömmlinge der noch unbekannten Wasserstoffverbindung CH3.C5N4H3 and nenne die letztere Methylpurin." (In order to make possible a rational nomenclature for the numerous existing substances, I regarded them as derivatives of a still unknown hydrogen compound, CH3.C5N4H3, and call the latter "methylpurine".) - ^ Fischer E (1898). "Ueber das Purin und seine Methylderivate" [On purine and its methyl derivatives] (PDF). Berichte der Deutschen Chemischen Gesellschaft. 31 (3): 2550–74. doi:10.1002/cber.18980310304. Retrieved 2016-04-20.

From p. 2550 Archived 2020-10-18 at the Wayback Machine: "…hielt ich es für zweckmäßig, alle diese Produkte ebenso wie die Harnsäure als Derivate der sauerstofffreien Verbindung C5H4N4 zu betrachten, und wählte für diese den Namen Purin, welcher aus den Wörtern purum und uricum kombiniert war." (…I regarded it as expedient to consider all of these products, just like uric acid, as derivatives of the oxygen-free compound C5H4N4, and chose for them the name "purine", which was formed from the words purum and uricum.) - Scheele CW (1776). "Examen chemicum calculi urinari" [A chemical examination of kidney stones]. Opuscula. 2: 73.
- Davies O, Mendes P, Smallbone K, Malys N (April 2012). "Characterisation of multiple substrate-specific (d)ITP/(d)XTPase and modelling of deaminated purine nucleotide metabolism". BMB Reports. 45 (4): 259–264. doi:10.5483/BMBRep.2012.45.4.259. PMID 22531138.
- ^ Brown, Anne M.; Hoopes, Samantha L.; White, Robert H.; Sarisky, Catherine A. (2011). "Purine biosynthesis in archaea: Variations on a theme". Biology Direct. 6: 63. doi:10.1186/1745-6150-6-63. PMC 3261824. PMID 22168471.
- Bernstein, H. (1961). "Imidazole Compounds Accumulated by Purine Mutants of Neurospora crassa". Journal of General Microbiology. 25: 41–46. doi:10.1099/00221287-25-1-41.
- Choi HK, Atkinson K, Karlson EW, Willett W, Curhan G (March 2004). "Purine-rich foods, dairy and protein intake, and the risk of gout in men". The New England Journal of Medicine. 350 (11): 1093–1103. doi:10.1056/NEJMoa035700. PMID 15014182.
- Yamada H, Okamoto T (1972). "A One-step Synthesis of Purine Ring from Formamide". Chemical & Pharmaceutical Bulletin. 20 (3): 623. doi:10.1248/cpb.20.623. Archived from the original on 2016-05-16.
- Saladino R, Crestini C, Ciciriello F, Costanzo G, Di Mauro E (December 2006). "About a formamide-based origin of informational polymers: syntheses of nucleobases and favourable thermodynamic niches for early polymers". Origins of Life and Evolution of the Biosphere. 36 (5–6): 523–531. Bibcode:2006OLEB...36..523S. doi:10.1007/s11084-006-9053-2. PMID 17136429. S2CID 36278915.
- , "Process for preparing adenine", issued 1966-11-10 Archived 2021-05-26 at the Wayback Machine
- Sanchez RA, Ferris JP, Orgel LE (December 1967). "Studies in prebiotic synthesis. II. Synthesis of purine precursors and amino acids from aqueous hydrogen cyanide". Journal of Molecular Biology. 30 (2): 223–253. doi:10.1016/S0022-2836(67)80037-8. PMID 4297187.
- Ferris JP, Orgel LE (March 1966). "An Unusual Photochemical Rearrangement in the Synthesis of Adenine from Hydrogen Cyanide". Journal of the American Chemical Society. 88 (5): 1074. doi:10.1021/ja00957a050.
- Ferris JP, Kuder JE, Catalano AW (November 1969). "Photochemical reactions and the chemical evolution of purines and nicotinamide derivatives". Science. 166 (3906): 765–6. Bibcode:1969Sci...166..765F. doi:10.1126/science.166.3906.765. PMID 4241847. S2CID 695243.
- Oro J, Kamat SS (April 1961). "Amino-acid synthesis from hydrogen cyanide under possible primitive earth conditions". Nature. 190 (4774): 442–3. Bibcode:1961Natur.190..442O. doi:10.1038/190442a0. PMID 13731262. S2CID 4219284.
- Bauer W (1985). Houben-Weyl Methods of Organic Chemistry. Vol. E 5 (4th Supplement ed.). Thieme Georg Verlag. p. 1547. ISBN 978-3-13-181154-7.
- Hassner A, Stumer C (2002). Organic Syntheses Based on Name Reactions (2nd ed.). Elsevier. ISBN 0-08-043259-X.
- Nam I, Nam HG, Zare RN (January 2018). "Abiotic synthesis of purine and pyrimidine ribonucleosides in aqueous microdroplets". Proc Natl Acad Sci U S A. 115 (1): 36–40. doi:10.1073/pnas.1718559115. PMC 5776833. PMID 29255025.
- Becker S, Thoma I, Deutsch A, Gehrke T, Mayer P, Zipse H, Carell T (May 2016). "A high-yielding, strictly regioselective prebiotic purine nucleoside formation pathway". Science. 352 (6287): 833–6. doi:10.1126/science.aad2808. PMID 27174989.
External links
| Nucleic acid constituents | |||||||
|---|---|---|---|---|---|---|---|
| Nucleobase | |||||||
| Nucleoside |
| ||||||
| Nucleotide (Nucleoside monophosphate) |
| ||||||
| Nucleoside diphosphate | |||||||
| Nucleoside triphosphate | |||||||