| Revision as of 07:11, 22 November 2006 edit203.198.195.13 (talk)No edit summary← Previous edit | Latest revision as of 03:19, 21 November 2024 edit undoMy very best wishes (talk | contribs)Extended confirmed users56,376 editsNo edit summary | ||
| (441 intermediate revisions by more than 100 users not shown) | |||
| Line 1: | Line 1: | ||
| {{short description|Chemical changes in proteins following their translation from mRNA}} | |||
| '''Posttranslational modification''' is the ] modification of a ] after its ]. It is one of the later steps in ] for many proteins. | |||
| ]. At the top, the ribosome translates a mRNA sequence into a protein, insulin, and passes the protein through the ], where it is cut, folded, and held in shape by disulfide (-S-S-) bonds. Then the protein passes through the ], where it is packaged into a vesicle. In the vesicle, more parts are cut off, and it turns into mature insulin.]] | |||
| ] of insulin, as described.]] | |||
| A protein (also called a ]) is a chain of ]s. During protein synthesis, 20 different amino acids can be incorporated in proteins. After translation, the posttranslational modification (PTM) of amino acids extends the range of functions of the protein by attaching to it other biochemical ]s such as ], ], various ]s and ]s, by changing the chemical nature of an amino acid (e.g. ]) or by making structural changes, like the formation of ]s. | |||
| In ], '''post-translational modification''' ('''PTM''') is the ] process of changing ]s following ]. PTMs may involve ] or occur spontaneously. Proteins are created by ], which ] ] into ]s, which may then change to form the mature protein product. PTMs are important components in cell ], as for example when ]s are converted to ]s. | |||
| Also, ]s may remove amino acids from the ] of the protein, or cut the peptide chain in the middle. For instance, the peptide ] ] is cut twice after disulfide bonds are formed, and a ] is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bonds. | |||
| Post-translational modifications can occur on the ] ]s or at the protein's ] or ] termini.<ref>{{cite book|last1=Pratt|first1=Charlotte W.|authorlink1=Charlotte W. Pratt|authorlink2=Judith G. Voet|authorlink3=Donald Voet|first2=Judith G.|last2=Voet|last3=Voet|first3=Donald|url=https://books.google.com/books?id=h0FCAQAAIAAJ|title=Fundamentals of Biochemistry: Life at the Molecular Level|date=2006|publisher=Wiley|location=Hoboken, NJ|oclc=1280801548|isbn=9780471214953|archive-date=13 July 2012|archive-url=https://archive.org/details/fundamentalsofbi00voet_0|edition=2nd }}</ref> They can expand the chemical set of the 22 ] by changing an existing ] or adding a new one such as phosphate. ] is highly effective for controlling the enzyme activity and is the most common change after translation. <ref name="khoury">{{cite journal|vauthors=Khoury GA, Baliban RC, Floudas CA|author-link3=Christodoulos Floudas|date=September 2011|title=Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database|journal=]|volume=1|pages=90|bibcode=2011NatSR...1E..90K|doi=10.1038/srep00090|pmc=3201773|pmid=22034591}}</ref> Many ] and ] proteins also have ] molecules attached to them in a process called ], which can promote ] and improve stability as well as serving regulatory functions. Attachment of ] molecules, known as ], often targets a protein or part of a protein attached to the ]. | |||
| Other modifications, like ], are part of common mechanisms for controlling the behavior of a protein, for instance activating or inactivating an enzyme. | |||
| Other forms of post-translational modification consist of cleaving ]s, as in processing a ] to a mature form or removing the initiator ] residue. The formation of ]s from ] residues may also be referred to as a post-translational modification.<ref name=lodish>{{cite book|vauthors=Lodish H, Berk A, Zipursky SL|display-authors=etal|chapter=17.6, Post-Translational Modifications and Quality Control in the Rough ER|title=Molecular Cell Biology|date=2000|publisher=W. H. Freeman|location=New York|isbn=978-0-7167-3136-8|edition=4th|chapter-url=https://www.ncbi.nlm.nih.gov/books/NBK21741/|url=https://archive.org/details/molecularcellbio00lodi}}</ref> For instance, the peptide ] ] is cut twice after disulfide bonds are formed, and a ] is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bonds. | |||
| Some types of post-translational modification are consequences of ]. ] is one example that targets the modified protein for degradation and can result in the formation of protein aggregates.<ref>{{cite journal|vauthors=Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A|year=2006|title=Protein carbonylation, cellular dysfunction, and disease progression|journal=]|volume=10|issue=2|pages=389–406|doi=10.1111/j.1582-4934.2006.tb00407.x|pmc=3933129|pmid=16796807}}</ref><ref>{{cite journal|vauthors=Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA|date=August 2008|title=Oxidative stress and covalent modification of protein with bioactive aldehydes|journal=]|volume=283|issue=32|pages=21837–41|doi=10.1074/jbc.R700019200|pmc=2494933|pmid=18445586|doi-access=free}}</ref> Specific amino acid modifications can be used as ]s indicating oxidative damage.<ref>{{cite journal | vauthors = Gianazza E, Crawford J, Miller I | title = Detecting oxidative post-translational modifications in proteins | journal = Amino Acids | volume = 33 | issue = 1 | pages = 51–6 | date = July 2007 | pmid = 17021655 | doi = 10.1007/s00726-006-0410-2 | s2cid = 23819101 }}</ref> | |||
| Sites that often undergo post-translational modification are those that have a functional group that can serve as a ] in the reaction: the ] groups of ], ], and ]; the ] forms of ], ], and ]; the ] ] of ]; the ]s of ] and ]; and the N- and C-termini. In addition, although the ] of ] is a weak nucleophile, it can serve as an attachment point for ]s. Rarer modifications can occur at oxidized ]s and at some ]s in side chains.<ref name=walsh>{{cite book|last1=Walsh|first1=Christopher T.|title=Posttranslational modification of proteins : expanding nature's inventory|date=2006|publisher=Roberts and Co. Publ.|location=Englewood|isbn=9780974707730}} {{rp|12–14}}</ref> | |||
| Post-translational modification of proteins can be experimentally detected by a variety of techniques, including ], ], and ]. | |||
| == PTMs involving addition of functional groups == | == PTMs involving addition of functional groups == | ||
| <!-- removed from display. See Talk ] Catalog (2004-5) p.263</ref> showing the amino acid residues as target of modification.]]--> | |||
| ===Addition by an enzyme ''in vivo''=== | |||
| ====Hydrophobic groups for membrane localization==== | |||
| * ] (a type of ]), attachment of ], a C<sub>14</sub> saturated acid | |||
| * ] (a type of acylation), attachment of ], a C<sub>16</sub> saturated acid | |||
| * ] or ], the addition of an ] group (e.g. ] and ]) | |||
| ** ] | |||
| ** ] | |||
| * ], ] formation via an amide bond to C-terminal tail | |||
| ====Cofactors for enhanced enzymatic activity==== | |||
| PTMs involving addition include: | |||
| * ] (a type of acylation), attachment of a ] (C<sub>8</sub>) functional group | |||
| * ], the addition of an ] group, usually at the ] of the protein | |||
| * ] moiety (] or ]) may be covalently attached | |||
| * ], the addition of an ] group (e.g. methyl, ethyl) | |||
| * ] attachment via ] bonds with ]s | |||
| ** ] the addition of a ] group, usually at ] or ] residues. (This is a type of alkylation.) | |||
| * ], acylation of conserved ] residues with a biotin appendage | |||
| * ], covalent linkage of ] residues to tubulin and some other proteins. | |||
| * ], covalent linkage of one to more than 40 ] residues to the ] C-terminal tail | |||
| * ], the addition of a ] group to either ], ], ], or ], resulting in a ] | |||
| * ], the addition of an ] group (e.g. farnesol and geranylgeraniol) | |||
| * ], attachment of a lipoate functionality | |||
| * ], the addition of a 4'-phosphopantetheinyl moiety from ], as in fatty acid, polyketide, non-ribosomal peptide and leucine biosynthesis | * ], the addition of a 4'-phosphopantetheinyl moiety from ], as in fatty acid, polyketide, non-ribosomal peptide and leucine biosynthesis | ||
| * ] ] formation | |||
| * ], the addition of a ] group, usually to ], ], ] or ] | |||
| * ], the addition of a sulfate group to a ]. | |||
| *Selenation | |||
| *C-terminal amidation | |||
| ====Modifications of translation factors==== | |||
| == PTMs involving addition of other proteins or peptides == | |||
| * ] formation (on a histidine found in ]) | |||
| * ISGylation, the ] linkage to the ISG15 protein (Interferon-Stimulated Gene 15) (2) | |||
| * ] attachment (on glutamate found in ])<ref name="pmid2569467">{{cite journal | vauthors = Whiteheart SW, Shenbagamurthi P, Chen L, Cotter RJ, Hart GW | title = Murine elongation factor 1 alpha (EF-1 alpha) is posttranslationally modified by novel amide-linked ethanolamine-phosphoglycerol moieties. Addition of ethanolamine-phosphoglycerol to specific glutamic acid residues on EF-1 alpha | journal = The Journal of Biological Chemistry | volume = 264 | issue = 24 | pages = 14334–41 | date = August 1989 | doi = 10.1016/S0021-9258(18)71682-7 | pmid = 2569467 | display-authors = etal | doi-access = free }}</ref> | |||
| * ], the ] linkage to the ] (Small Ubiquitin-related MOdifier) (1) | |||
| * ] formation (on conserved lysine of ] (eukaryotic) and ] (archaeal)) | |||
| * ], the ] linkage to the protein ubiquitin. | |||
| * ] addition on a conserved lysine of the ] (EFP) in most bacteria.<ref>{{cite journal | vauthors = Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M | title = The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine | journal = Nature Chemical Biology | volume = 7 | issue = 10 | pages = 667–9 | date = August 2011 | pmid = 21841797 | pmc = 3177975 | doi = 10.1038/nchembio.632 }}</ref> EFP is a homolog to ] (eukaryotic) and ] (archaeal) (see above). | |||
| == |
====Smaller chemical groups==== | ||
| * ], e.g. ''O''-acylation (]), ''N''-acylation (]), ''S''-acylation (]) | |||
| ** ], the addition of an ] group, either at the ] of the protein or at ] residues.<ref name="pmid29405707">{{cite journal | vauthors = Ali I, Conrad RJ, Verdin E, Ott M | title = Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics | journal = Chem Rev | volume = 118 | issue = 3 | pages = 1216–1252 | date = February 2018 | pmid = 29405707 | pmc = 6609103 | doi = 10.1021/acs.chemrev.7b00181 }}</ref> The reverse is called ]. | |||
| ** ] | |||
| * ], the addition of an ] group, e.g. ], ] | |||
| ** ] the addition of a ] group, usually at ] or ] residues. The reverse is called ]. | |||
| * <span id="amidation">amidation</span> at C-terminus. Formed by oxidative dissociation of a C-terminal Gly residue.<ref name="amidation">{{cite journal | vauthors = Bradbury AF, Smyth DG | title = Peptide amidation | journal = Trends in Biochemical Sciences | volume = 16 | issue = 3 | pages = 112–5 | date = March 1991 | pmid = 2057999 | doi = 10.1016/0968-0004(91)90044-v }}</ref> | |||
| * ] bond formation | |||
| ** ] addition | |||
| *** ], a ]-mediation addition | |||
| *** ], covalent linkage of ] residues to the N-terminus of tubulin and some other proteins.<ref name="pmid1967194">{{cite journal | vauthors = Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P | title = Posttranslational glutamylation of alpha-tubulin | journal = Science | volume = 247 | issue = 4938 | pages = 83–5 | date = January 1990 | pmid = 1967194 | doi = 10.1126/science.1967194 | bibcode = 1990Sci...247...83E }}</ref> (See ]) | |||
| *** ], covalent linkage of one to more than 40 ] residues to the ] C-terminal tail | |||
| * ] | |||
| * ] dependent on ]<ref>{{cite journal | vauthors = Walker CS, Shetty RP, Clark K, Kazuko SG, Letsou A, Olivera BM, Bandyopadhyay PK | title = On a potential global role for vitamin K-dependent gamma-carboxylation in animal systems. Evidence for a gamma-glutamyl carboxylase in Drosophila | journal = The Journal of Biological Chemistry | volume = 276 | issue = 11 | pages = 7769–74 | date = March 2001 | pmid = 11110799 | doi = 10.1074/jbc.M009576200 | display-authors = etal | doi-access = free }}</ref> | |||
| * ], the addition of a ] group to either ], ], ], ], ], ], ], or ] resulting in a ]. Distinct from ], which is regarded as a nonenzymatic attachment of sugars. | |||
| ** ], addition of ''N''-acetylglucosamine to serine or threonine residues in a β-glycosidic linkage | |||
| ** polysialylation, addition of ], PSA, to ] | |||
| * ] | |||
| * ]: addition of an oxygen atom to the side-chain of a Pro or Lys residue | |||
| * ]: addition of an iodine atom to the aromatic ring of a tyrosine residue (e.g. in ]) | |||
| * ] such as ] | |||
| * ] (''O''-linked) or ] (''N''-linked) formation | |||
| ** ], the addition of a ] group, usually to ], ], and ] (''O''-linked), or ] (''N''-linked) | |||
| ** ], the addition of an ] moiety, usually to ] (''O''-linked), or ] and ] (''N''-linked) | |||
| ** uridylylation, the addition of an uridylyl-group (i.e. ], UMP), usually to tyrosine | |||
| * ] | |||
| * ] formation | |||
| * ] | |||
| * ] | |||
| * ''S''-sulfenylation (''aka'' ''S''-sulphenylation), reversible covalent addition of one oxygen atom to the ] group of a ] residue<ref name="CysOx">{{cite journal | vauthors = Chung HS, Wang SB, Venkatraman V, Murray CI, Van Eyk JE | title = Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system | journal = Circulation Research | volume = 112 | issue = 2 | pages = 382–92 | date = January 2013 | pmid = 23329793 | pmc = 4340704 | doi = 10.1161/CIRCRESAHA.112.268680 | display-authors = 1 }}</ref> | |||
| <!-- old ref{{Cite journal|title = MDD–SOH: exploiting maximal dependence decomposition to identify S-sulfenylation sites with substrate motifs|date = 26 September 2015|journal = Bioinformatics|doi = 10.1093/bioinformatics/btv558|pmid = 26411868|vauthors = Bui VM, Lu CT, Ho TT, Lee TY|volume = 32|issue = 2|pages = 165–72}}--> | |||
| * ''S''-sulfinylation, normally irreversible covalent addition of two oxygen atoms to the ] group of a ] residue<ref name="CysOx" /> | |||
| * ''S''-sulfonylation, normally irreversible covalent addition of three oxygen atoms to the ] group of a ] residue, resulting in the formation of a ] residue<ref name="CysOx" /> | |||
| * ] addition of a ] group to ] | |||
| * ], the addition of a sulfate group to a ]. | |||
| ===Non-enzymatic modifications ''in vivo''=== | |||
| * ], or '''deimination''' the conversion of ] to ] | |||
| Examples of non-enzymatic PTMs are glycation, glycoxidation, nitrosylation, oxidation, succination, and lipoxidation.<ref>"The Advanced Lipoxidation End-Product Malondialdehyde-Lysine in Aging and Longevity" PMID 33203089 PMC7696601</ref> | |||
| * ], the conversion of ] to ] or ] to ] | |||
| * ], the addition of a sugar molecule to a protein without the controlling action of an enzyme. | |||
| * ] the addition of ] to a protein's N-terminus or the side-chain of Lys.<ref name="pmid21768218">{{cite journal | vauthors = Jaisson S, Pietrement C, Gillery P | title = Carbamylation-derived products: bioactive compounds and potential biomarkers in chronic renal failure and atherosclerosis | journal = Clinical Chemistry | volume = 57 | issue = 11 | pages = 1499–505 | date = November 2011 | pmid = 21768218 | doi = 10.1373/clinchem.2011.163188 | doi-access = free }}</ref> | |||
| * ] the addition of carbon monoxide to other organic/inorganic compounds. | |||
| * spontaneous ] formation, as found in many surface proteins of ].<ref name="pmid21055949">{{cite journal | vauthors = Kang HJ, Baker EN | title = Intramolecular isopeptide bonds: protein crosslinks built for stress? | journal = Trends in Biochemical Sciences | volume = 36 | issue = 4 | pages = 229–37 | date = April 2011 | pmid = 21055949 | doi = 10.1016/j.tibs.2010.09.007 }}</ref> | |||
| ===Non-enzymatic additions ''in vitro''=== | |||
| == PTMs involving structural changes == | |||
| * ]: covalent attachment of a biotin moiety using a biotinylation reagent, typically for the purpose of labeling a protein. | |||
| * carbamylation: the addition of Isocyanic acid to a protein's N-terminus or the side-chain of Lys or Cys residues, typically resulting from exposure to urea solutions.<ref name="stark_1960">{{cite journal |vauthors=Stark GR, Stein WH, Moore X | title = Reactions of the Cyanate Present in Aqueous Urea with Amino Acids and Proteins | journal = J Biol Chem | volume = 235 | issue = 11 | pages = 3177–3181 | year = 1960 | doi = 10.1016/S0021-9258(20)81332-5 | doi-access = free }}</ref> | |||
| * oxidation: addition of one or more Oxygen atoms to a susceptible side-chain, principally of Met, Trp, His or Cys residues. Formation of ] bonds between Cys residues. | |||
| * ]: covalent attachment of ] (PEG) using a pegylation reagent, typically to the N-terminus or the side-chains of Lys residues. Pegylation is used to improve the efficacy of protein pharmaceuticals. | |||
| ==Conjugation with other proteins or peptides== | |||
| * ]ation, the ] linkage to the protein ubiquitin. | |||
| * ], the covalent linkage to the ] (Small Ubiquitin-related MOdifier)<ref>Van G. Wilson (Ed.) (2004). {{webarchive|url=https://web.archive.org/web/20050209075122/http://horizonpress.com/hsp/books/sumo.html |date=2005-02-09 }}. Horizon Bioscience. {{ISBN|0-9545232-8-8}}.</ref> | |||
| * ], the covalent linkage to the ] | |||
| * ISGylation, the covalent linkage to the ] protein (Interferon-Stimulated Gene 15)<ref>{{cite journal | vauthors = Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, Peterson LF, Shuai K, Zhang DE | title = Protein ISGylation modulates the JAK-STAT signaling pathway | journal = Genes & Development | volume = 17 | issue = 4 | pages = 455–60 | date = February 2003 | pmid = 12600939 | pmc = 195994 | doi = 10.1101/gad.1056303 }}</ref> | |||
| * ], the covalent linkage to the ] | |||
| == Chemical modification of amino acids == | |||
| * ], or '''deimination''', the conversion of ] to ]<ref>{{cite journal | vauthors = Klareskog L, Rönnelid J, Lundberg K, Padyukov L, Alfredsson L | title = Immunity to citrullinated proteins in rheumatoid arthritis | journal = Annual Review of Immunology | volume = 26 | pages = 651–75 | year = 2008 | pmid = 18173373 | doi = 10.1146/annurev.immunol.26.021607.090244 | url = https://zenodo.org/record/894124 }}</ref> | |||
| * ], the conversion of ] to ] or ] to ] | |||
| * ], the conversion to an ] by ] of ] and ], or ] of ] and ]<ref>{{cite journal | vauthors = Brennan DF, Barford D | title = Eliminylation: a post-translational modification catalyzed by phosphothreonine lyases | journal = Trends in Biochemical Sciences | volume = 34 | issue = 3 | pages = 108–14 | date = March 2009 | pmid = 19233656 | doi = 10.1016/j.tibs.2008.11.005 }}</ref> | |||
| == Structural changes == | |||
| * ]s, the covalent linkage of two ] amino acids | * ]s, the covalent linkage of two ] amino acids | ||
| * ]s, the covalent linkage of 1 lysine and 1 or 2 cystine residues via an oxygen atom (NOS and SONOS bridges)<ref>{{cite journal |last1=Rabe von Pappenheim |first1=Fabian |last2=Wensien |first2=Marie |last3=Ye |first3=Jin |last4=Uranga |first4=Jon |last5=Irisarri |first5=Iker |last6=de Vries |first6=Jan |last7=Funk |first7=Lisa-Marie |last8=Mata |first8=Ricardo A. |last9=Tittmann |first9=Kai |title=Widespread occurrence of covalent lysine–cysteine redox switches in proteins |journal=Nature Chemical Biology |date=April 2022 |volume=18 |issue=4 |pages=368–375 |doi=10.1038/s41589-021-00966-5 |doi-access=free|pmid=35165445 |pmc=8964421 }}</ref> | |||
| * ], cleavage of a protein at a peptide bond | |||
| * ], cleavage of a protein at a peptide bond | |||
| * ] formation, via the cyclisation of asparagine or aspartic acid amino-acid residues | |||
| * ] | |||
| ** of ] by ] | |||
| ** of ] in ], a frog ] | |||
| ** of ] in ], also a frog opioid peptide | |||
| * ], self-catalytic removal of ]s analogous to mRNA processing | |||
| == |
== Statistics == | ||
| ===Common PTMs by frequency=== | |||
| In 2011, statistics of each post-translational modification experimentally and putatively detected have been compiled using proteome-wide information from the Swiss-Prot database.<ref>{{cite journal | vauthors = Khoury GA, Baliban RC, Floudas CA | title = Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database | journal = Scientific Reports | volume = 1 | issue = 90 | pages = 90 | date = September 2011 | pmid = 22034591 | pmc = 3201773 | doi = 10.1038/srep00090 | bibcode = 2011NatSR...1E..90K }}</ref> The 10 most common experimentally found modifications were as follows:<ref>{{cite web|url=http://selene.princeton.edu/PTMCuration/|title=Proteome-Wide Post-Translational Modification Statistics|website=selene.princeton.edu|access-date=2011-07-22|archive-url=https://web.archive.org/web/20120830234041/http://selene.princeton.edu/PTMCuration/|archive-date=2012-08-30|url-status=dead}}</ref> | |||
| {| class="wikitable" | |||
| !Frequency | |||
| !Modification | |||
| |- | |||
| |58383 | |||
| |] | |||
| |- | |||
| |6751 | |||
| |] | |||
| |- | |||
| |5526 | |||
| |] | |||
| |- | |||
| |2844 | |||
| |] | |||
| |- | |||
| |1619 | |||
| |] | |||
| |- | |||
| |1523 | |||
| |] | |||
| |- | |||
| |1133 | |||
| |] | |||
| |- | |||
| |878 | |||
| |] | |||
| |- | |||
| |826 | |||
| |] | |||
| |- | |||
| |504 | |||
| |] | |||
| |} | |||
| === Common PTMs by residue === | |||
| * cleavage and formation of disulfide bridges during the production of ] | |||
| Some common post-translational modifications to specific amino-acid residues are shown below. Modifications occur on the side-chain unless indicated otherwise. | |||
| * PTM of ]s as regulation of transcription: ] | |||
| * PTM of RNA polymerase II as regulation of transcription: ] | |||
| {| class="wikitable sortable" style="text-align: left" | |||
| == References == | |||
| |- | |||
| ! Amino Acid !! Abbrev. !! Modification | |||
| |- | |||
| ! ] | |||
| | Ala or A | |||
| | ] (N-terminus) | |||
| |- | |||
| ! ] | |||
| | Arg or R | |||
| | deimination to ], ] | |||
| |- | |||
| ! ] | |||
| | Asn or N | |||
| | ] to Asp or iso(Asp), ], ] | |||
| |- | |||
| ! ] | |||
| | Asp or D | |||
| | ] to isoaspartic acid, ] | |||
| |- | |||
| ! ] | |||
| | Cys or C | |||
| | ]-bond formation, oxidation to sulfenic, sulfinic or sulfonic acid, ], ] (N-terminus), ] | |||
| |- | |||
| ! ] | |||
| | Gln or Q | |||
| | cyclization to ] (N-terminus), deamidation to ] or ] formation to a ] by a ] | |||
| |- | |||
| ! ] | |||
| | Glu or E | |||
| | cyclization to ] (N-terminus), ] | |||
| |- | |||
| ! ] | |||
| | Gly or G | |||
| | N-] (N-terminus), ] (N-terminus) | |||
| |- | |||
| ! ] | |||
| | His or H | |||
| | ] | |||
| |- | |||
| ! ] | |||
| | Ile or I | |||
| | | |||
| |- | |||
| ! ] | |||
| | Leu or L | |||
| | | |||
| |- | |||
| ! ] | |||
| | Lys or K | |||
| | ], ], ], ], ] leading to ], ] | |||
| |- | |||
| ! ] | |||
| | Met or M | |||
| | ] (N-terminus), N-linked Ubiquitination, oxidation to sulfoxide or sulfone | |||
| |- | |||
| ! ] | |||
| | Phe or F | |||
| | | |||
| |- | |||
| ! ] | |||
| | Pro or P | |||
| | ] | |||
| |- | |||
| ! ] | |||
| | Ser or S | |||
| | ], ], ] (N-terminus) | |||
| |- | |||
| ! ] | |||
| | Thr or T | |||
| | ], ], ] (N-terminus) | |||
| |- | |||
| ! ] | |||
| | Trp or W | |||
| | mono- or di-oxidation, formation of ], ] | |||
| |- | |||
| ! ] | |||
| | Tyr or Y | |||
| | ], phosphorylation | |||
| |- | |||
| ! ] | |||
| | Val or V | |||
| | ] (N-terminus) | |||
| |} | |||
| == Databases and tools == | |||
| 1. Van G. Wilson (Ed.) (2004). . Horizon Bioscience. ISBN 0-9545232-8-8. | |||
| ] | |||
| 2. Malakhova, Oxana A.; Yan, Ming; Malakhov, Michael P.; Yuan, Youzhong; Ritchie, Kenneth J.; Kim, Keun Il; Peterson, Luke F.; Shuai, Ke; and Dong-Er Zhang. (2003). . ''Genes & Development'' '''17''' (4), 455-460. | |||
| Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features. | |||
| === List of resources === | |||
| * <ref>{{cite journal | vauthors = Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E | title = PhosphoSitePlus, 2014: mutations, PTMs and recalibrations | journal = Nucleic Acids Research | volume = 43 | issue = Database issue | pages = D512-20 | date = January 2015 | pmid = 25514926 | pmc = 4383998 | doi = 10.1093/nar/gku1267 }}</ref> – A database of comprehensive information and tools for the study of mammalian protein post-translational modification | |||
| * ]<ref name="ReferenceB">{{cite journal | vauthors = Goel R, Harsha HC, Pandey A, Prasad TS | title = Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis | journal = Molecular BioSystems | volume = 8 | issue = 2 | pages = 453–63 | date = February 2012 | pmid = 22159132 | pmc = 3804167 | doi = 10.1039/c1mb05340j }}</ref> – A database of proteins and post-translational modifications experimentally | |||
| * ]<ref name="ReferenceB"/> – A database for different modifications and understand different proteins, their class, and function/process related to disease causing proteins | |||
| * ]<ref>{{cite journal | vauthors = Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N | title = PROSITE, a protein domain database for functional characterization and annotation | journal = Nucleic Acids Research | volume = 38 | issue = Database issue | pages = D161-6 | date = January 2010 | pmid = 19858104 | pmc = 2808866 | doi = 10.1093/nar/gkp885 }}</ref> – A database of Consensus patterns for many types of PTM's including sites | |||
| * ]<ref>{{cite journal | vauthors = Garavelli JS | title = The RESID Database of Protein Modifications: 2003 developments | journal = Nucleic Acids Research | volume = 31 | issue = 1 | pages = 499–501 | date = January 2003 | pmid = 12520062 | pmc = 165485 | doi = 10.1093/nar/gkg038 }}</ref> – A database consisting of a collection of annotations and structures for PTMs. | |||
| * <ref>{{cite journal | vauthors = Huang H, Arighi CN, Ross KE, Ren J, Li G, Chen SC, Wang Q, Cowart J, Vijay-Shanker K, Wu CH | title = iPTMnet: an integrated resource for protein post-translational modification network discovery | journal = Nucleic Acids Research | volume = 46 | issue = 1 | pages = D542–D550 | date = January 2018 | pmid = 2914561 | pmc = 5753337 | doi = 10.1093/nar/gkx1104}}</ref>– A database that integrates PTM information from several knowledgbases and text mining results. | |||
| * ]<ref name="ReferenceA"/> – A database that shows different PTM's and information regarding their chemical components/structures and a frequency for amino acid modified site | |||
| * has PTM information although that may be less comprehensive than in more specialized databases.] | |||
| * <ref>{{cite journal | vauthors = Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J, Kahsay R, Olivier-Van Stichelen S | title = The human ''O''-GlcNAcome database and meta-analysis | journal = Scientific Data | volume = 8 | date = January 2021 | issue = 1 | page = 25 | pmid = 33479245 | pmc = 7820439 | doi = 10.1038/s41597-021-00810-4 | bibcode = 2021NatSD...8...25W }}</ref><ref>{{cite journal | vauthors = Malard F, Wulff-Fuentes E, Berendt RR, Didier G, Olivier-Van Stichelen S | title = Automatization and self-maintenance of the O-GlcNAcome catalog: a smart scientific database | journal = Database (Oxford) | volume = 2021 | date = July 2021 | page = 1 | pmid = 34279596 | doi = 10.1093/database/baab039 | pmc = 8288053 }}</ref> - A curated database for protein O-GlcNAcylation and referencing more than 14 000 protein entries and 10 000 ''O''-GlcNAc sites. | |||
| === Tools === | |||
| List of software for visualization of proteins and their PTMs | |||
| * ]<ref>{{cite journal | vauthors = Warnecke A, Sandalova T, Achour A, Harris RA | title = PyTMs: a useful PyMOL plugin for modeling common post-translational modifications | journal = BMC Bioinformatics | volume = 15 | issue = 1 | pages = 370 | date = November 2014 | pmid = 25431162 | pmc = 4256751 | doi = 10.1186/s12859-014-0370-6 | doi-access = free }}</ref> – introduce a set of common PTM's into protein models | |||
| * ]<ref>{{cite journal | vauthors = Yang Y, Peng X, Ying P, Tian J, Li J, Ke J, Zhu Y, Gong Y, Zou D, Yang N, Wang X, Mei S, Zhong R, Gong J, Chang J, Miao X | title = AWESOME: a database of SNPs that affect protein post-translational modifications | journal = Nucleic Acids Research | volume = 47 | issue = D1 | pages = D874–D880 | date = January 2019 | pmid = 30215764 | pmc = 6324025 | doi = 10.1093/nar/gky821 }}</ref> – Interactive tool to see the role of single nucleotide polymorphisms to PTM's | |||
| * ]<ref>{{cite journal | vauthors = Morris JH, Huang CC, Babbitt PC, Ferrin TE | title = structureViz: linking Cytoscape and UCSF Chimera | journal = Bioinformatics | volume = 23 | issue = 17 | pages = 2345–7 | date = September 2007 | pmid = 17623706 | doi = 10.1093/bioinformatics/btm329 | doi-access = free }}</ref> – Interactive Database to visualize molecules | |||
| == Case examples == | |||
| {{more citations needed section|date=January 2016}} | |||
| * Cleavage and formation of ]s during the production of ] | |||
| * PTM of ]s as regulation of ]: ] | |||
| * PTM of ] as regulation of transcription | |||
| * Cleavage of polypeptide chains as crucial for lectin specificity<ref>{{cite web|url=http://www.proteopedia.org/index.php/1tp8|title=1tp8 - Proteopedia, life in 3D|website=www.proteopedia.org}}</ref> | |||
| ==See also== | |||
| * ] | |||
| * ] | |||
| ==References== | |||
| {{reflist|30em}} | |||
| ==External links== | |||
| * in ] | |||
| * | |||
| * — from the ] database | |||
| * | |||
| {{Protein topics}} | |||
| {{Protein primary structure}} | {{Protein primary structure}} | ||
| {{Posttranslational modification}} | |||
| {{Gene expression}} | |||
| {{Portal bar|Biology}} | |||
| {{DEFAULTSORT:Posttranslational Modification}} | |||
| ] | |||
| ] | |||
| ] | ] | ||
| ] | ] | ||
| ] | ] | ||
| ] | |||
| ] | |||
| ] | |||
Latest revision as of 03:19, 21 November 2024
Chemical changes in proteins following their translation from mRNA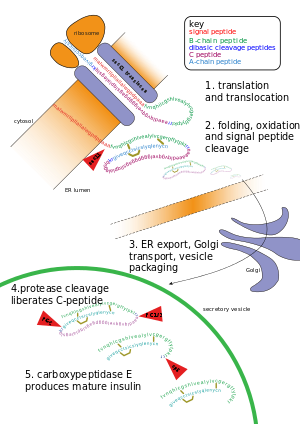
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.
Post-translational modifications can occur on the amino acid side chains or at the protein's C- or N- termini. They can expand the chemical set of the 22 amino acids by changing an existing functional group or adding a new one such as phosphate. Phosphorylation is highly effective for controlling the enzyme activity and is the most common change after translation. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a process called glycosylation, which can promote protein folding and improve stability as well as serving regulatory functions. Attachment of lipid molecules, known as lipidation, often targets a protein or part of a protein attached to the cell membrane.
Other forms of post-translational modification consist of cleaving peptide bonds, as in processing a propeptide to a mature form or removing the initiator methionine residue. The formation of disulfide bonds from cysteine residues may also be referred to as a post-translational modification. For instance, the peptide hormone insulin is cut twice after disulfide bonds are formed, and a propeptide is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bonds.
Some types of post-translational modification are consequences of oxidative stress. Carbonylation is one example that targets the modified protein for degradation and can result in the formation of protein aggregates. Specific amino acid modifications can be used as biomarkers indicating oxidative damage.
Sites that often undergo post-translational modification are those that have a functional group that can serve as a nucleophile in the reaction: the hydroxyl groups of serine, threonine, and tyrosine; the amine forms of lysine, arginine, and histidine; the thiolate anion of cysteine; the carboxylates of aspartate and glutamate; and the N- and C-termini. In addition, although the amide of asparagine is a weak nucleophile, it can serve as an attachment point for glycans. Rarer modifications can occur at oxidized methionines and at some methylene groups in side chains.
Post-translational modification of proteins can be experimentally detected by a variety of techniques, including mass spectrometry, Eastern blotting, and Western blotting.
PTMs involving addition of functional groups
Addition by an enzyme in vivo
Hydrophobic groups for membrane localization
- myristoylation (a type of acylation), attachment of myristate, a C14 saturated acid
- palmitoylation (a type of acylation), attachment of palmitate, a C16 saturated acid
- isoprenylation or prenylation, the addition of an isoprenoid group (e.g. farnesol and geranylgeraniol)
- glypiation, glycosylphosphatidylinositol (GPI) anchor formation via an amide bond to C-terminal tail
Cofactors for enhanced enzymatic activity
- lipoylation (a type of acylation), attachment of a lipoate (C8) functional group
- flavin moiety (FMN or FAD) may be covalently attached
- heme C attachment via thioether bonds with cysteines
- phosphopantetheinylation, the addition of a 4'-phosphopantetheinyl moiety from coenzyme A, as in fatty acid, polyketide, non-ribosomal peptide and leucine biosynthesis
- retinylidene Schiff base formation
Modifications of translation factors
- diphthamide formation (on a histidine found in eEF2)
- ethanolamine phosphoglycerol attachment (on glutamate found in eEF1α)
- hypusine formation (on conserved lysine of eIF5A (eukaryotic) and aIF5A (archaeal))
- beta-Lysine addition on a conserved lysine of the elongation factor P (EFP) in most bacteria. EFP is a homolog to eIF5A (eukaryotic) and aIF5A (archaeal) (see above).
Smaller chemical groups
- acylation, e.g. O-acylation (esters), N-acylation (amides), S-acylation (thioesters)
- acetylation, the addition of an acetyl group, either at the N-terminus of the protein or at lysine residues. The reverse is called deacetylation.
- formylation
- alkylation, the addition of an alkyl group, e.g. methyl, ethyl
- methylation the addition of a methyl group, usually at lysine or arginine residues. The reverse is called demethylation.
- amidation at C-terminus. Formed by oxidative dissociation of a C-terminal Gly residue.
- amide bond formation
- amino acid addition
- arginylation, a tRNA-mediation addition
- polyglutamylation, covalent linkage of glutamic acid residues to the N-terminus of tubulin and some other proteins. (See tubulin polyglutamylase)
- polyglycylation, covalent linkage of one to more than 40 glycine residues to the tubulin C-terminal tail
- amino acid addition
- butyrylation
- gamma-carboxylation dependent on Vitamin K
- glycosylation, the addition of a glycosyl group to either arginine, asparagine, cysteine, hydroxylysine, serine, threonine, tyrosine, or tryptophan resulting in a glycoprotein. Distinct from glycation, which is regarded as a nonenzymatic attachment of sugars.
- O-GlcNAc, addition of N-acetylglucosamine to serine or threonine residues in a β-glycosidic linkage
- polysialylation, addition of polysialic acid, PSA, to NCAM
- malonylation
- hydroxylation: addition of an oxygen atom to the side-chain of a Pro or Lys residue
- iodination: addition of an iodine atom to the aromatic ring of a tyrosine residue (e.g. in thyroglobulin)
- nucleotide addition such as ADP-ribosylation
- phosphate ester (O-linked) or phosphoramidate (N-linked) formation
- phosphorylation, the addition of a phosphate group, usually to serine, threonine, and tyrosine (O-linked), or histidine (N-linked)
- adenylylation, the addition of an adenylyl moiety, usually to tyrosine (O-linked), or histidine and lysine (N-linked)
- uridylylation, the addition of an uridylyl-group (i.e. uridine monophosphate, UMP), usually to tyrosine
- propionylation
- pyroglutamate formation
- S-glutathionylation
- S-nitrosylation
- S-sulfenylation (aka S-sulphenylation), reversible covalent addition of one oxygen atom to the thiol group of a cysteine residue
- S-sulfinylation, normally irreversible covalent addition of two oxygen atoms to the thiol group of a cysteine residue
- S-sulfonylation, normally irreversible covalent addition of three oxygen atoms to the thiol group of a cysteine residue, resulting in the formation of a cysteic acid residue
- succinylation addition of a succinyl group to lysine
- sulfation, the addition of a sulfate group to a tyrosine.
Non-enzymatic modifications in vivo
Examples of non-enzymatic PTMs are glycation, glycoxidation, nitrosylation, oxidation, succination, and lipoxidation.
- glycation, the addition of a sugar molecule to a protein without the controlling action of an enzyme.
- carbamylation the addition of Isocyanic acid to a protein's N-terminus or the side-chain of Lys.
- carbonylation the addition of carbon monoxide to other organic/inorganic compounds.
- spontaneous isopeptide bond formation, as found in many surface proteins of Gram-positive bacteria.
Non-enzymatic additions in vitro
- biotinylation: covalent attachment of a biotin moiety using a biotinylation reagent, typically for the purpose of labeling a protein.
- carbamylation: the addition of Isocyanic acid to a protein's N-terminus or the side-chain of Lys or Cys residues, typically resulting from exposure to urea solutions.
- oxidation: addition of one or more Oxygen atoms to a susceptible side-chain, principally of Met, Trp, His or Cys residues. Formation of disulfide bonds between Cys residues.
- pegylation: covalent attachment of polyethylene glycol (PEG) using a pegylation reagent, typically to the N-terminus or the side-chains of Lys residues. Pegylation is used to improve the efficacy of protein pharmaceuticals.
Conjugation with other proteins or peptides
- ubiquitination, the covalent linkage to the protein ubiquitin.
- SUMOylation, the covalent linkage to the SUMO protein (Small Ubiquitin-related MOdifier)
- neddylation, the covalent linkage to the Nedd protein
- ISGylation, the covalent linkage to the ISG15 protein (Interferon-Stimulated Gene 15)
- pupylation, the covalent linkage to the prokaryotic ubiquitin-like protein
Chemical modification of amino acids
- citrullination, or deimination, the conversion of arginine to citrulline
- deamidation, the conversion of glutamine to glutamic acid or asparagine to aspartic acid
- eliminylation, the conversion to an alkene by beta-elimination of phosphothreonine and phosphoserine, or dehydration of threonine and serine
Structural changes
- disulfide bridges, the covalent linkage of two cysteine amino acids
- lysine-cysteine bridges, the covalent linkage of 1 lysine and 1 or 2 cystine residues via an oxygen atom (NOS and SONOS bridges)
- proteolytic cleavage, cleavage of a protein at a peptide bond
- isoaspartate formation, via the cyclisation of asparagine or aspartic acid amino-acid residues
- racemization
- of serine by protein-serine epimerase
- of alanine in dermorphin, a frog opioid peptide
- of methionine in deltorphin, also a frog opioid peptide
- protein splicing, self-catalytic removal of inteins analogous to mRNA processing
Statistics
Common PTMs by frequency
In 2011, statistics of each post-translational modification experimentally and putatively detected have been compiled using proteome-wide information from the Swiss-Prot database. The 10 most common experimentally found modifications were as follows:
| Frequency | Modification |
|---|---|
| 58383 | Phosphorylation |
| 6751 | Acetylation |
| 5526 | N-linked glycosylation |
| 2844 | Amidation |
| 1619 | Hydroxylation |
| 1523 | Methylation |
| 1133 | O-linked glycosylation |
| 878 | Ubiquitylation |
| 826 | Pyrrolidone carboxylic acid |
| 504 | Sulfation |
Common PTMs by residue
Some common post-translational modifications to specific amino-acid residues are shown below. Modifications occur on the side-chain unless indicated otherwise.
Databases and tools

Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features.
List of resources
- PhosphoSitePlus – A database of comprehensive information and tools for the study of mammalian protein post-translational modification
- ProteomeScout – A database of proteins and post-translational modifications experimentally
- Human Protein Reference Database – A database for different modifications and understand different proteins, their class, and function/process related to disease causing proteins
- PROSITE – A database of Consensus patterns for many types of PTM's including sites
- RESID – A database consisting of a collection of annotations and structures for PTMs.
- iPTMnet – A database that integrates PTM information from several knowledgbases and text mining results.
- dbPTM – A database that shows different PTM's and information regarding their chemical components/structures and a frequency for amino acid modified site
- Uniprot has PTM information although that may be less comprehensive than in more specialized databases.

Effect of PTMs on protein function and physiological processes. - The O-GlcNAc Database - A curated database for protein O-GlcNAcylation and referencing more than 14 000 protein entries and 10 000 O-GlcNAc sites.
Tools
List of software for visualization of proteins and their PTMs
- PyMOL – introduce a set of common PTM's into protein models
- AWESOME – Interactive tool to see the role of single nucleotide polymorphisms to PTM's
- Chimera – Interactive Database to visualize molecules
Case examples
| This section needs additional citations for verification. Please help improve this article by adding citations to reliable sources in this section. Unsourced material may be challenged and removed. (January 2016) (Learn how and when to remove this message) |
- Cleavage and formation of disulfide bridges during the production of insulin
- PTM of histones as regulation of transcription: RNA polymerase control by chromatin structure
- PTM of RNA polymerase II as regulation of transcription
- Cleavage of polypeptide chains as crucial for lectin specificity
See also
References
- Pratt, Charlotte W.; Voet, Judith G.; Voet, Donald (2006). Fundamentals of Biochemistry: Life at the Molecular Level (2nd ed.). Hoboken, NJ: Wiley. ISBN 9780471214953. OCLC 1280801548. Archived from the original on 13 July 2012.
- Khoury GA, Baliban RC, Floudas CA (September 2011). "Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database". Scientific Reports. 1: 90. Bibcode:2011NatSR...1E..90K. doi:10.1038/srep00090. PMC 3201773. PMID 22034591.
- Lodish H, Berk A, Zipursky SL, et al. (2000). "17.6, Post-Translational Modifications and Quality Control in the Rough ER". Molecular Cell Biology (4th ed.). New York: W. H. Freeman. ISBN 978-0-7167-3136-8.
- Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A (2006). "Protein carbonylation, cellular dysfunction, and disease progression". Journal of Cellular and Molecular Medicine. 10 (2): 389–406. doi:10.1111/j.1582-4934.2006.tb00407.x. PMC 3933129. PMID 16796807.
- Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA (August 2008). "Oxidative stress and covalent modification of protein with bioactive aldehydes". The Journal of Biological Chemistry. 283 (32): 21837–41. doi:10.1074/jbc.R700019200. PMC 2494933. PMID 18445586.
- Gianazza E, Crawford J, Miller I (July 2007). "Detecting oxidative post-translational modifications in proteins". Amino Acids. 33 (1): 51–6. doi:10.1007/s00726-006-0410-2. PMID 17021655. S2CID 23819101.
- Walsh, Christopher T. (2006). Posttranslational modification of proteins : expanding nature's inventory. Englewood: Roberts and Co. Publ. ISBN 9780974707730.
- Whiteheart SW, Shenbagamurthi P, Chen L, Cotter RJ, Hart GW, et al. (August 1989). "Murine elongation factor 1 alpha (EF-1 alpha) is posttranslationally modified by novel amide-linked ethanolamine-phosphoglycerol moieties. Addition of ethanolamine-phosphoglycerol to specific glutamic acid residues on EF-1 alpha". The Journal of Biological Chemistry. 264 (24): 14334–41. doi:10.1016/S0021-9258(18)71682-7. PMID 2569467.
- Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M (August 2011). "The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine". Nature Chemical Biology. 7 (10): 667–9. doi:10.1038/nchembio.632. PMC 3177975. PMID 21841797.
- Ali I, Conrad RJ, Verdin E, Ott M (February 2018). "Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics". Chem Rev. 118 (3): 1216–1252. doi:10.1021/acs.chemrev.7b00181. PMC 6609103. PMID 29405707.
- Bradbury AF, Smyth DG (March 1991). "Peptide amidation". Trends in Biochemical Sciences. 16 (3): 112–5. doi:10.1016/0968-0004(91)90044-v. PMID 2057999.
- Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P (January 1990). "Posttranslational glutamylation of alpha-tubulin". Science. 247 (4938): 83–5. Bibcode:1990Sci...247...83E. doi:10.1126/science.1967194. PMID 1967194.
- Walker CS, Shetty RP, Clark K, Kazuko SG, Letsou A, Olivera BM, Bandyopadhyay PK, et al. (March 2001). "On a potential global role for vitamin K-dependent gamma-carboxylation in animal systems. Evidence for a gamma-glutamyl carboxylase in Drosophila". The Journal of Biological Chemistry. 276 (11): 7769–74. doi:10.1074/jbc.M009576200. PMID 11110799.
- ^ Chung HS, et al. (January 2013). "Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system". Circulation Research. 112 (2): 382–92. doi:10.1161/CIRCRESAHA.112.268680. PMC 4340704. PMID 23329793.
- "The Advanced Lipoxidation End-Product Malondialdehyde-Lysine in Aging and Longevity" PMID 33203089 PMC7696601
- Jaisson S, Pietrement C, Gillery P (November 2011). "Carbamylation-derived products: bioactive compounds and potential biomarkers in chronic renal failure and atherosclerosis". Clinical Chemistry. 57 (11): 1499–505. doi:10.1373/clinchem.2011.163188. PMID 21768218.
- Kang HJ, Baker EN (April 2011). "Intramolecular isopeptide bonds: protein crosslinks built for stress?". Trends in Biochemical Sciences. 36 (4): 229–37. doi:10.1016/j.tibs.2010.09.007. PMID 21055949.
- Stark GR, Stein WH, Moore X (1960). "Reactions of the Cyanate Present in Aqueous Urea with Amino Acids and Proteins". J Biol Chem. 235 (11): 3177–3181. doi:10.1016/S0021-9258(20)81332-5.
- Van G. Wilson (Ed.) (2004). Sumoylation: Molecular Biology and Biochemistry Archived 2005-02-09 at the Wayback Machine. Horizon Bioscience. ISBN 0-9545232-8-8.
- Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, Peterson LF, Shuai K, Zhang DE (February 2003). "Protein ISGylation modulates the JAK-STAT signaling pathway". Genes & Development. 17 (4): 455–60. doi:10.1101/gad.1056303. PMC 195994. PMID 12600939.
- Klareskog L, Rönnelid J, Lundberg K, Padyukov L, Alfredsson L (2008). "Immunity to citrullinated proteins in rheumatoid arthritis". Annual Review of Immunology. 26: 651–75. doi:10.1146/annurev.immunol.26.021607.090244. PMID 18173373.
- Brennan DF, Barford D (March 2009). "Eliminylation: a post-translational modification catalyzed by phosphothreonine lyases". Trends in Biochemical Sciences. 34 (3): 108–14. doi:10.1016/j.tibs.2008.11.005. PMID 19233656.
- Rabe von Pappenheim, Fabian; Wensien, Marie; Ye, Jin; Uranga, Jon; Irisarri, Iker; de Vries, Jan; Funk, Lisa-Marie; Mata, Ricardo A.; Tittmann, Kai (April 2022). "Widespread occurrence of covalent lysine–cysteine redox switches in proteins". Nature Chemical Biology. 18 (4): 368–375. doi:10.1038/s41589-021-00966-5. PMC 8964421. PMID 35165445.
- Khoury GA, Baliban RC, Floudas CA (September 2011). "Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database". Scientific Reports. 1 (90): 90. Bibcode:2011NatSR...1E..90K. doi:10.1038/srep00090. PMC 3201773. PMID 22034591.
- "Proteome-Wide Post-Translational Modification Statistics". selene.princeton.edu. Archived from the original on 2012-08-30. Retrieved 2011-07-22.
- ^ Lee TY, Huang HD, Hung JH, Huang HY, Yang YS, Wang TH (January 2006). "dbPTM: an information repository of protein post-translational modification". Nucleic Acids Research. 34 (Database issue): D622-7. doi:10.1093/nar/gkj083. PMC 1347446. PMID 16381945.
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E (January 2015). "PhosphoSitePlus, 2014: mutations, PTMs and recalibrations". Nucleic Acids Research. 43 (Database issue): D512-20. doi:10.1093/nar/gku1267. PMC 4383998. PMID 25514926.
- ^ Goel R, Harsha HC, Pandey A, Prasad TS (February 2012). "Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis". Molecular BioSystems. 8 (2): 453–63. doi:10.1039/c1mb05340j. PMC 3804167. PMID 22159132.
- Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N (January 2010). "PROSITE, a protein domain database for functional characterization and annotation". Nucleic Acids Research. 38 (Database issue): D161-6. doi:10.1093/nar/gkp885. PMC 2808866. PMID 19858104.
- Garavelli JS (January 2003). "The RESID Database of Protein Modifications: 2003 developments". Nucleic Acids Research. 31 (1): 499–501. doi:10.1093/nar/gkg038. PMC 165485. PMID 12520062.
- Huang H, Arighi CN, Ross KE, Ren J, Li G, Chen SC, Wang Q, Cowart J, Vijay-Shanker K, Wu CH (January 2018). "iPTMnet: an integrated resource for protein post-translational modification network discovery". Nucleic Acids Research. 46 (1): D542–D550. doi:10.1093/nar/gkx1104. PMC 5753337. PMID 2914561.
- Audagnotto M, Dal Peraro M (2017-03-31). "In silico prediction tools and molecular modeling". Computational and Structural Biotechnology Journal. 15: 307–319. doi:10.1016/j.csbj.2017.03.004. PMC 5397102. PMID 28458782.
- Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J, Kahsay R, Olivier-Van Stichelen S (January 2021). "The human O-GlcNAcome database and meta-analysis". Scientific Data. 8 (1): 25. Bibcode:2021NatSD...8...25W. doi:10.1038/s41597-021-00810-4. PMC 7820439. PMID 33479245.
- Malard F, Wulff-Fuentes E, Berendt RR, Didier G, Olivier-Van Stichelen S (July 2021). "Automatization and self-maintenance of the O-GlcNAcome catalog: a smart scientific database". Database (Oxford). 2021: 1. doi:10.1093/database/baab039. PMC 8288053. PMID 34279596.
- Warnecke A, Sandalova T, Achour A, Harris RA (November 2014). "PyTMs: a useful PyMOL plugin for modeling common post-translational modifications". BMC Bioinformatics. 15 (1): 370. doi:10.1186/s12859-014-0370-6. PMC 4256751. PMID 25431162.
- Yang Y, Peng X, Ying P, Tian J, Li J, Ke J, Zhu Y, Gong Y, Zou D, Yang N, Wang X, Mei S, Zhong R, Gong J, Chang J, Miao X (January 2019). "AWESOME: a database of SNPs that affect protein post-translational modifications". Nucleic Acids Research. 47 (D1): D874–D880. doi:10.1093/nar/gky821. PMC 6324025. PMID 30215764.
- Morris JH, Huang CC, Babbitt PC, Ferrin TE (September 2007). "structureViz: linking Cytoscape and UCSF Chimera". Bioinformatics. 23 (17): 2345–7. doi:10.1093/bioinformatics/btm329. PMID 17623706.
- "1tp8 - Proteopedia, life in 3D". www.proteopedia.org.
External links
- Controlled vocabulary of post-translational modifications in Uniprot
- List of posttranslational modifications in ExPASy
- Browse SCOP domains by PTM — from the dcGO database
- Overview and description of commonly used post-translational modification detection techniques
| Proteins | |
|---|---|
| Processes | |
| Structures | |
| Types | |
| Protein primary structure and posttranslational modifications | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| General | |||||||||||||||||||||||||
| N terminus | |||||||||||||||||||||||||
| C terminus | |||||||||||||||||||||||||
| Single specific AAs |
| ||||||||||||||||||||||||
| Crosslinks between two AAs |
| ||||||||||||||||||||||||
| Crosslinks between three AAs | |||||||||||||||||||||||||
| Crosslinks between four AAs |
| ||||||||||||||||||||||||
| Posttranslational modification | |||||
|---|---|---|---|---|---|
| Chaperones/ protein folding |
| ||||
| Protein targeting | |||||
| Ubiquitin (ubiquitylation) | |||||
| Ubiquitin-like proteins (UBL) |
| ||||
| Gene expression | |||||||
|---|---|---|---|---|---|---|---|
| Introduction to genetics | |||||||
| Transcription |
| ||||||
| Translation |
| ||||||
| Regulation | |||||||
| Influential people | |||||||